Extended Data Fig. 3. Genetic context of octocoral TCs.
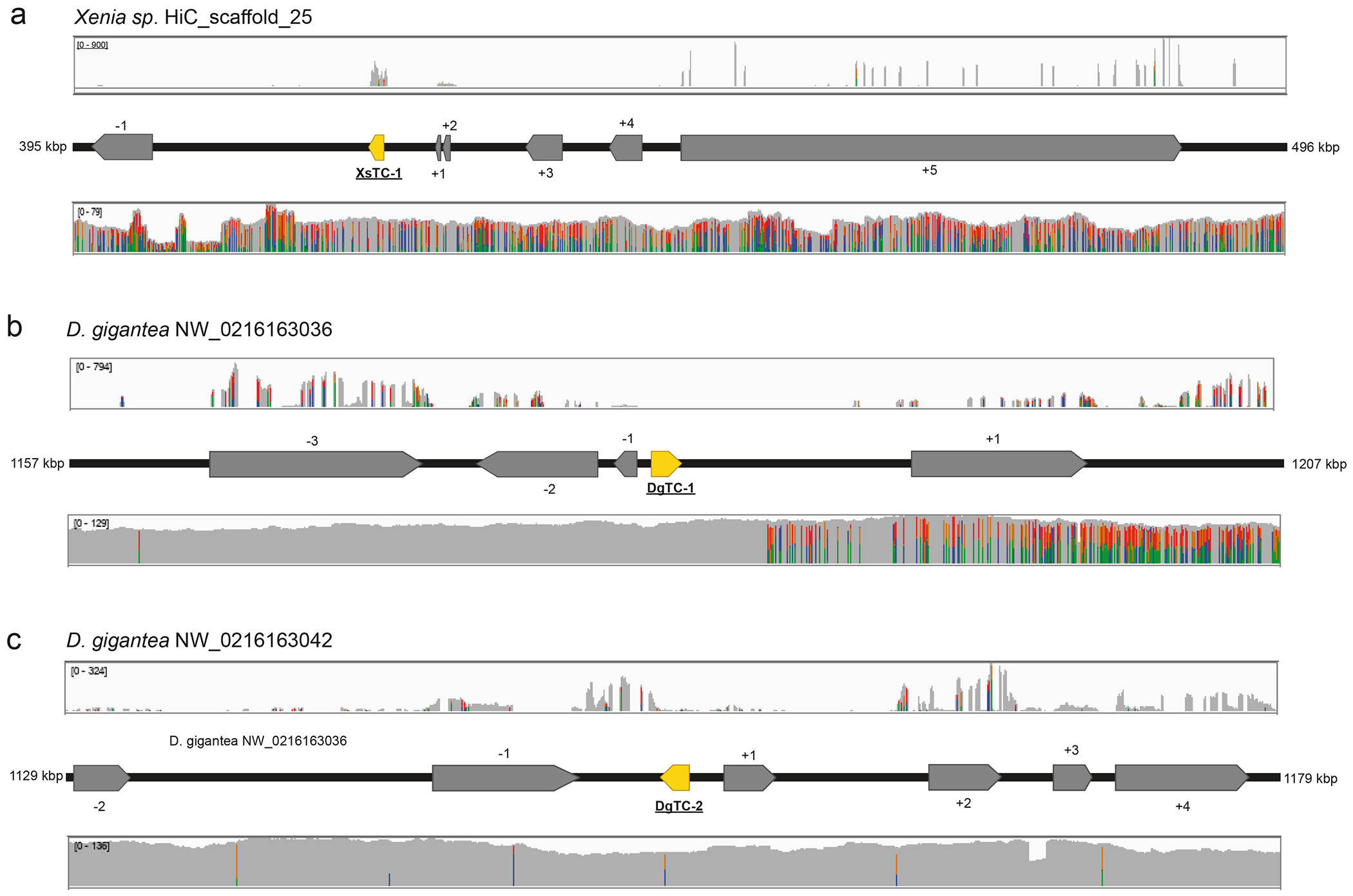
Mapped transcripts (top), gene annotations (middle) and mapped long-reads (bottom) for the indicated regions are shown. a) Context of XsTC-1 from Xenia sp. b) Context of DgTC-1 from D. gigantea. c) Context of DgTC-2 from D. gigantea. All genes are coded on long eukaryotic contigs (see Supplementary Table 4) with continuous long read coverage between 60 and 130 (Oxford Nanopore for a, PacBio for b and c). While the expression of DgTC-1 and DgTC-2 is low, XsTC-1 is highly expressed. For gene IDs see Supplementary Tables 5–7. The co-occurrence of two different bases in the same position (marked by colors) results from the heterozygosity of D. gigantea and Xenia sp.
