Abstract
Several reports demonstrated that SARS-CoV-2 variant-of-concern B.1.1.529 (Omicron) exhibits a high degree of escape from antibody neutralization. Therefore, it is critical to determine how well the second line of adaptive immunity, T cell memory, performs against Omicron. To that purpose, we analyzed a human cohort (n=327 subjects) of two- or three- dose messenger RNA (mRNA) vaccine recipients and COVID-19 post infection subjects. We report that T cell responses against Omicron were largely preserved. IFN-γ producing T cell responses remained equivalent to the response against the ancestral strain (WA1/2020), with some (~20%) loss in IL-2 single or IL-2+IFNγ+ poly-functional responses. Three-dose vaccinated participants had similar responses to Omicron relative to post COVID-19 participants and exhibited responses significantly higher than those receiving two mRNA vaccine doses. These results provide further evidence that a three-dose vaccine regimen benefits the induction of optimal functional T cell immune memory.
Introduction
The B.1.1.529 (Omicron) SARS-CoV-2 variant was first reported to the World Health Organization (WHO) by the South African government in 2021(1) and was subsequently designated variant of concern (VOC). Omicron has accumulated a large number of mutations compared to the original Wuhan strain; a total of 60 out of which 50 were nonsynonymus(2). 32 of those mutations reside in the spike protein, which is the sole target of the majority of SARS-CoV-2 vaccines. This raised concerns about the potential for increased transmissibility and escape from vaccine protection. Considerable evidence points to increased transmissibility of Omicron as compared to previous VOCs, including the highly transmissible B.1.617.2 (Delta) variant. A cohort analysis of household transmission in England showed that two-fold more Omicron index cases gave rise to a secondary household case in comparison to Delta(3). Omicron is now the dominant SARS-CoV-2 variant in many parts of the world(4, 5). Early studies demonstrated a substantial reduction in antibody neutralizing capacity against Omicron(6, 7) which was maintained best in sera from individuals who experienced infection and were subsequently vaccinated(8). However, the spread of Omicron in South Africa has been followed by a decrease in hospitalizations(9) and death rates(10). Given the large degree of escape from antibody responses, T cell mediated immunity could be essential to prevent Omicron-induced severe COVID-19. Depletion of T cells in convalescent macaques resulted in impaired immunity against rechallenge with SARS-CoV-2, suggesting a significant role for T cells in the context of subprotective antibody titres(11). Human clinical studies also demonstrated a link between SARS-CoV-2 T cell responses and reduced disease severity(12, 13). Thus, it is paramount to determine how well T cell immunity against Omicron is preserved. We and others have shown that T cell immunity against Delta, Gamma and other variants was largely preserved in mRNA vaccine recipients(14, 15). However, earlier variants contained drastically lower numbers of mutations in the spike protein. To assess T cell immune memory to the Omicron variant we have analyzed a large cohort of participants recruited into the CDC-sponsored AZ HEROES research study(16). Participants in this cohort included subjects sampled post SARS-CoV-2 infection; fully vaccinated subjects (2 doses of mRNA vaccine); subjects post infection receiving two doses of mRNA vaccine; participants receiving three doses of mRNA vaccine; and pre-pandemic controls. We have measured polyfunctional T cell responses in peripheral blood mononuclear cells (PBMCs) to overlapping peptide pools corresponding to SARS-CoV-2 S proteins of USA-WA1/2020 (ancestral strain) and the Omicron variant. We report that T cell responses to spike protein of the Omicron variant are largely preserved. We found no waning of T cell IFN-γ responses upon stimulation with Omicron peptide pools compared to original strain, with a slight reduction in IL-2-producing and polyfunctional IFN-γ+IL-2+ double-positive cells and IFN-γ+IL-2+GrB+ triple positive cells. Similar results were obtained in a recently published study by Tarke et al.(17). Additionally, we report that T cell responses in participants receiving two doses of mRNA vaccines were lower than post infection, post infection + 2 vaccine doses and in those receiving three vaccine doses, suggesting that three doses of mRNA should be considered as an optimal, full vaccination regimen to induce robust T cell immunity.
Materials and methods
Study participants
This study was approved by the University of Arizona IRB (protocol #2102460536 and #1410545697) and the Oregon Health and Science University IRB (protocol #00003007, pre-pandemic controls) and was conducted in accordance with all federal state and local regulations. Prepandemic samples included healthy community-dwelling individuals recruited in Tucson, Arizona and Portland, Oregon prior to December 2019. All COVID-19 positive participants tested positive by PCR before April 1st 2021 prior to emergence of Delta variant in Arizona(18). All subjects > 55 years of age were considered older adults and subjects younger than 50 were considered adult. We did not include subjects aged 50–55. Blood was drawn into BD Vacutainer Blood Collection Tubes with Sodium Heparin. PBMCs were separated by ficoll gradient separation and cryopreserved in fetal calf serum (FCS) + 10% DMSO.
FLUOROSpot assays
Cryopreserved PBMC (5 × 106/sample) were thawed in prewarmed RPMI-1640 media supplemented with L-glutamine (Lonza) + 10% FCS. Thawed PBMCS were rested for 3–4h at 37 °C in X-VIVO™-15 Medium (Lonza) supplemented with 5% human-AB serum. Cells were then stimulated with ~1 nmol of peptide pool corresponding to spike of US-WA (wild-type) or Omicron (B.1.1.529) variant (16-mer peptide pools, overlapping by 10 amino acids (21st century Biochemicals Inc.). Cell suspensions were transferred to pre-coated human IFN-γ, TNF-α, IL-2, Granzyme-B (GrB) FLUOROSpot kits (Mabtech, Inc.) and developed after 42 h according to manufacturer instructions. Spots were imaged and counted using Iris FLUOROspot reader (Mabtech).
Flow cytometry
1 ×106 PBMCs were stimulated with ~1 nmol of peptide pool corresponding to spike protein of US-WA (wild-type) or Omicron (B.1.1.529) variant (21st century Biochemicals Inc.) for 24h in X-VIVO™-15 Medium (Lonza) supplemented with 5% human-AB serum at 37 °C. Unstimulated wells from each participant were used for subtraction of background. Cells were stained with surface antibodies in PBS (Lonza) + 2% FCS for 1h, stained with the live dead fixable blue dye for 30 min (Thermofisher) and then fixed and permeabilized using the FoxP3 Fix/Perm kit (eBioscience). Samples were acquired using a Cytek Aurora cytometer and analyzed by FlowJo software (Tree Star).
Statistical analysis
Graph Pad Prism v9 was used for statistical analysis. Upon inspection of data distribution by Shapiro-Wilks normality test, differences between paired samples treated with different peptide pools were calculated by two-tailed Wilcoxon rank test. Differences between the subject groups were calculated by Kruskal Wallis test with Dunn’s post hoc correction and. For all figures data presented as mean ± standard error of the mean. For all statistical differences *p<0.05, **p<0.01, ***p<0.001. ****p<0.0001.
Results and Discussion
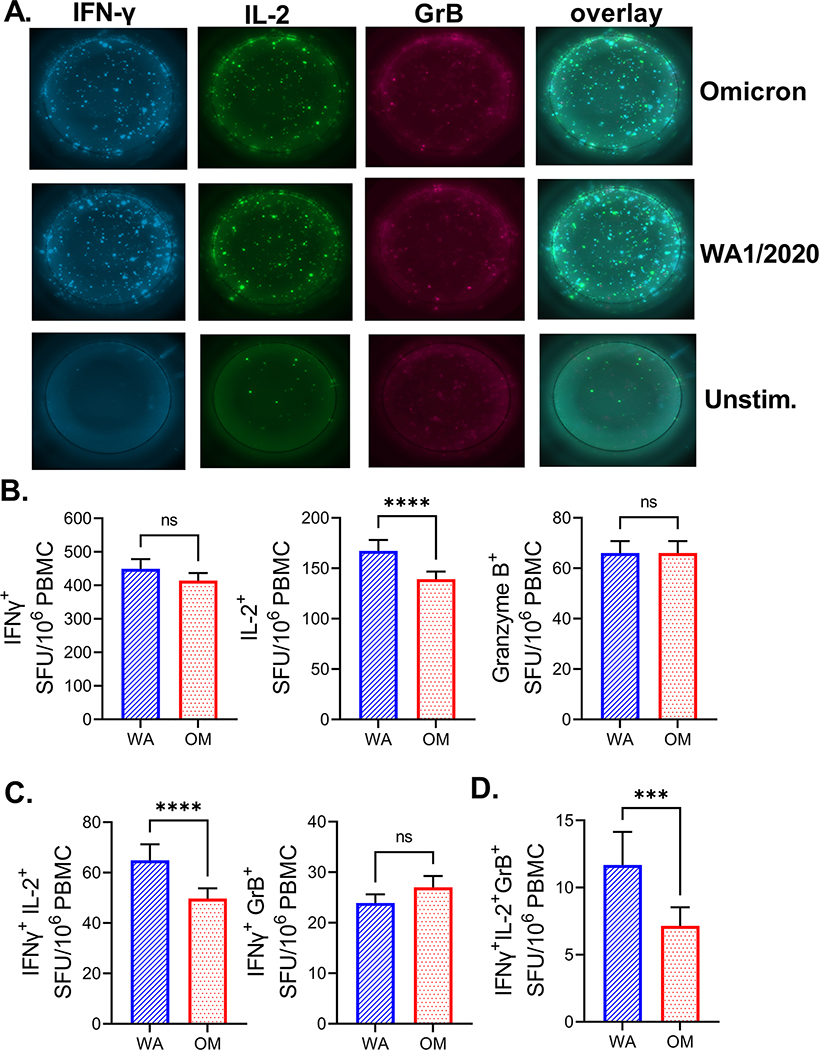
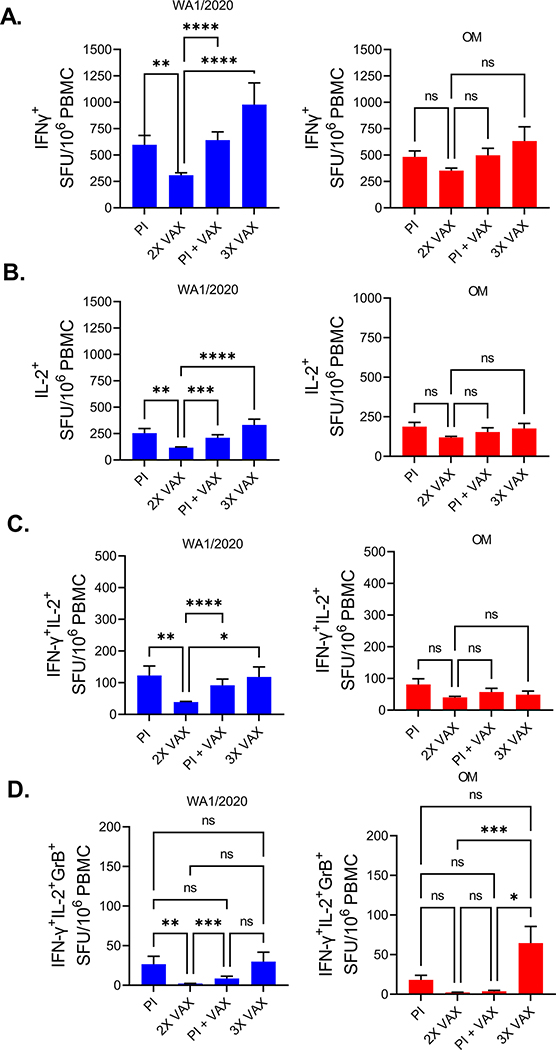
A total of 359 participants were included in this study: (i) 210 received two doses of SARS-CoV-2 mRNA vaccines (2X VAX); (ii) 54 were SARS-CoV-2 post infection samples with no additional vaccination (PI); (iii) 39 were vaccinated following SARS-CoV-2 infection (PI + VAX); (iv) 24 received three doses of mRNA vaccines (3X VAX); and (v) 32 participants recruited before 09/2019 (demographics in Supplemental Table 1). We have analyzed T cell responses of the participants to the spike protein of USA-WA1/2020 and Omicron variant by stimulating PBMCs with overlapping peptide pools spanning the spike protein and measuring production of T cell effector cytokines IFN-γ, IL-2 and Granzyme B (GrB) by FLUOROSpot. For each sample, the number of spot forming units (SFU) of each cytokine was calculated by subtracting the SFU number measured in the unstimulated wells from the SFU number of stimulated wells (representative FLUOROSpot images in Figure 1A). Spike peptide pool induced a 5-fold increase in IFN-γ SFU in immunized (vaccinated or post COVID-19) participants while no effect was evident in the pre-pandemic samples (Supplemental Figure 1). When comparing samples from all immunized participants, the number of IFN-γ SFU between Omicron and WA1/2020 peptide stimulated wells was equal (Figure 1B, left panel). However, the number of IL-2 SFU was reduced (Figure 1B, middle panel) around 16.8%. There was no difference in GrB SFU between the peptide pools (Figure 1B, right panel). Next, we analyzed the number of polyfunctional (double or triple cytokine positive) T cells. We detected a decrease in IFN-γ IL-2 double positive cells which were decreased 23.3% when stimulated with Omicron peptides, but not IFN-γ GrB double positive cells (Figure 1C), while triple positive (IFN-γ, IL-2, GrB) were 38.7% decreased with Omicron peptide pool stimulation (Figure 1D), however this was numerically the smallest population. Based on this large sample (N=327 exposed/immunized participants) we can conclude that the decrease in T cell immunity to mutated Omicron peptide is measurable, but the response is ~80% preserved. We next investigated the differences in T cell responses between the groups of study subjects. When stimulated with the WA1/2020 peptide pool, recipients of two doses of mRNA vaccine had significantly lower numbers of IFN-γ SFU (Figure 2A, left panel) than all other groups. A similar trend was measured with the Omicron peptide pool, albeit without reaching statistical significance (Figure 2A, right panel). Similarly, number of IL-2 SFU was decreased in the 2X VAX group stimulated with WA1/2020 peptides (Figure 2B). Double positive (IFN-γ+IL-2+) and triple positive (IFN-γ+IL-2+GrB+) SFU were also lower in the 2X VAX group (Figure 2C). Interestingly, the magnitude of polyfunctional triple positive cells to Omicron was the highest in the 3X VAX group (Figure 2D) although third dose of the vaccine did not increase the number of single or double positive cells reacting to Omicron pools (Figures 2A–C). Jointly, these results imply that two doses of mRNA vaccine induced sub-maximal T cell immunity, and that at least three doses are required to achieve the same level of antigen specific T cells found in post-infection subjects. We also compared T cell responses to each variant between subjects vaccinated with vaccines from different manufacturers (Pfizer and Moderna) and found that while there was no difference in IFN-γ T cell responses (Supplemental Fig. 2, left panel) within vaccine manufacturers, there was a higher number of IL-2+ SFU in samples from Moderna vaccinated subjects in response to either WA1/2020 or Omicron as compared to their Pfizer vaccinated counterparts (Supplemental Fig. 2, middle panel). The subjects grouped by vaccine manufacturer did not differ in the number of GrB SFU (Supplemental Fig. 2. right panel). Next, we investigated whether T cell responses were dependent on the age of the subject in our largest group (2X VAX). Given that IL-2 was most impacted by the mutations in Omicron peptides as compared to WA1/2020 (Fig. 1) we first analyzed the impact of age by IL-2+ SFU. We have detected a minimal, but measurable, negative correlation of IL-2 responses with age (r=−0.16, p=0.02). There was no impact of age for double positive (IFN-γ+IL-2+) SFU (r=−0.11, p=0.09), whereas triple positive (IFN-γ+IL-2+GrB+) SFU were negatively correlated to age of the subject (r=−0.24, p<0.0001). Overall, change in T cell responses with the age of the participant was minimal. Given that the 3X vaccinated group was on average 8 years older than 2X VAX (58 vs 50 years average age, Supplemental Table 1) but had higher T cell responses than 2X VAX (Figure 2A–2D) we conclude that age is not a major determinant in T cell responses to mRNA vaccines following administration of multiple vaccine doses.
Figure 1. Robust SARS-CoV-2 specific T cells responses in immunized individuals.
A) 106 PBMCs per well were stimulated with spike peptide pools from USA-WA1/2020) and Omicron cultured for 42h in pre-coated IFN-γ, IL-2 and GrB FLUOROSpot plates. Both peptide pools induced responses of all three cytokines compared to unstimulated wells. B) All immunized subjects were pooled together (N=327). Omicron peptide pool induced equal IFN-γ and GrB response compared to USA-WA1/2020 but lower IL-2. C) Number of polyfunctional IFN-γ+IL-2+ cells but not IFN-γ+GrB+ cells was reduced after stimulation with Omicron pool. D) Triple positive IFN-γ+IL-2+GrB+ were also reduced following stimulation with Omicron peptides. Two-tailed Wilcoxon rank test. Data presented as mean ± standard error of the mean. Data from one experiment.
Figure 2. SARS-CoV-2 T cell responses are lower in recipients of two doses of mRNA vaccines.
A) IFN-γ SFU were reduced in 2X VAX group compared to other groups when their PBMCs were stimulated with USA-WA1/2020 peptide pool (left panel), similar trend was observed with Omicron peptides but without statistical significance. B) IL-2 SFU were also reduced in the 2X vax group stimulated with USA-WA1/2020 peptide pool as were C) double positive IFN-γ+IL-2+ cells. D) number of triple positive IFN-γ+IL-2+GrB+ cells was highest in 3X VAX group. Kruskal-Wallis test with Dunn’s correction for multiple comparisons. Post infection (PI) n=54, Post infection and vaccinated (PI +VAX) n=39, two dose vaccinated (2X VAX) n=210, three dose vaccinated (3X VAX) n=24. Data presented as mean ± standard error of the mean. Data from one experiment.
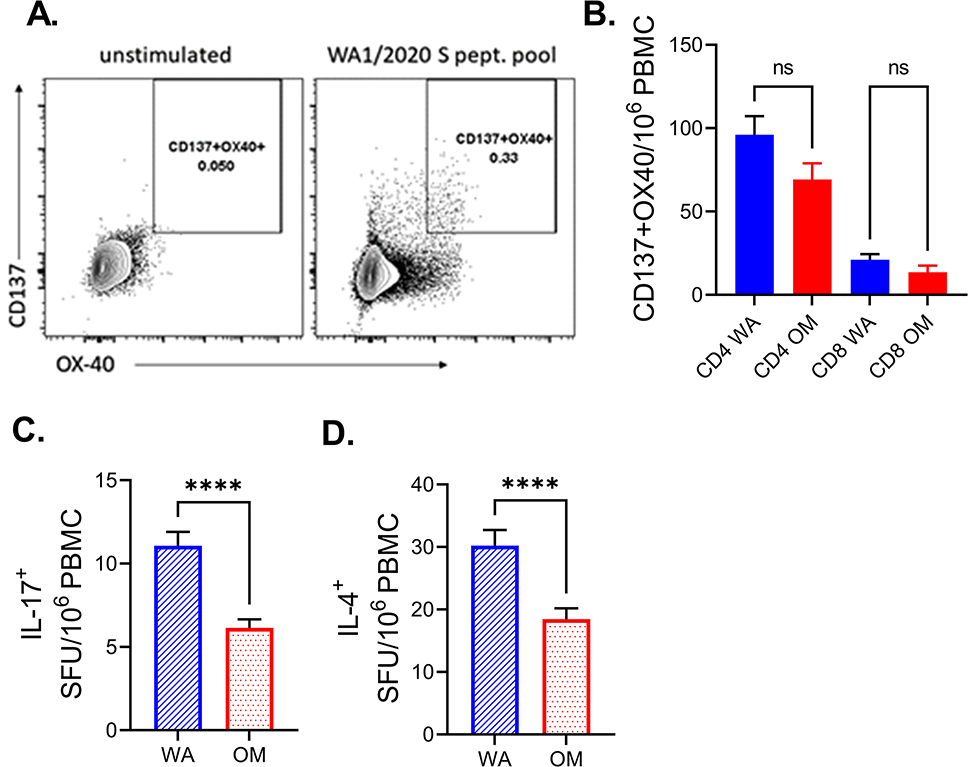
To further interrogate the phenotype of T cells responding to either WA1/2020 or Omicron we performed flow cytometry (FCM) on approximately half of the 2X VAX subject samples (randomly selected, n=96). Expression of costimulatory molecules CD137 (4-IBB) and OX-40 is one of the common methods for enumerating antigen (ag) specific T cells by flow cytometry, alongside intracellular staining for cytokines (19). Again, as with the FLUOROSpot assay, spike peptide pool induced a robust increase in CD137 and OX-40 activated T cells compared to stimulated wells (Supplemental Figure 3, representative FCM in Figure 3A). As previously reported(20), we have detected more CD4 helper T cells than cytotoxic CD8 T cells responding (Figure 3B). We did detect a slight decrease in the number of antigen specific CD4 and CD8 T cells in response to Omicron peptides vs. WA1/2020. However, the difference was not statistically significant (Figure 3B). Given that CD4+ helper T cells represent the bulk of the response we have additionally investigated the expression of Th polarizing cytokines IL-17a and IL-4 in the 2X VAX group. We found that both IL-17a (Figure 3C) and IL-4 (Figure 3D) were reduced in response to Omicron peptides, which could be interpreted as a positive finding, given that Th17 and Th2 responses would be expected to be counterproductive against viral infection.
Figure 3. Spike peptide pools induce a greater helper (CD4+) T cell response than cytotoxic (CD8+) T cell response.
A) representative flow cytometric plot of spike peptide induction of expression of costimulatory molecules CD137 and OX-40 on CD3+CD4+ T cells. B) Number of CD4+ cells expressing CD137 and OX-40 was higher than for CD8 T cells. Kruskal-Wallis test with Dunn’s correction for multiple comparisons C) IL-17a and D) IL-4 were reduced in response to Omicron peptides in subjects which received two doses of mRNA vaccines. Two-tailed Wilcoxon rank test. N=96. Data presented as mean ± standard error of the mean. Data from one experiment.
Overall, our findings highlight resilience of T cell responses generated in response to a pre-Omicron infection and/or to Wuhan-derived spike protein-based vaccines in the face of the B1.1.529 Omicron variant, particularly as measured by the IFN-γ production, which was the largest component of the antiviral T cell response. We did measure decreased IL-2 and polyfunctional responses, however these were generally reduced by less than 20–30%. Stratified analysis of subjects immunized by vaccines or prior infection clearly revealed that 2X VAX participants exhibited inferior responses to both Omicron and the ancestral strain relative to other groups, and in particular 3X VAX participants. Therefore, our results stress the need for a three-dose vaccine regimen to achieve robust T cell immunity.
Supplementary Material
Key points.
T cell responses against Omicron are well preserved post COVID-19 or mRNA vaccination
Recipients of two mRNA vaccine doses have lower T-cell responses
Acknowledgements
We would like to thank the personnel of UArizona Health Sciences Biobank for expert processing and storage of human blood samples. We would like to thank our CDC colleagues Drs Natalie J. Thornburg, Mark G. Thompson and Julie Mayo Lamberte for logistical help, input into study design and comments on the manuscript.
Supported in part by the USPHS award R37 AG020719 and the Bowman Professorship in Medical Sciences to J.N-Z. and the CDC HEROES project award 75D30120C08379 to J.L.B.
Data availability
The data and materials that support the findings of this study are available from the corresponding author upon request.
References
- 1.Karim SSA, and Karim QA 2021. Omicron SARS-CoV-2 variant: a new chapter in the COVID-19 pandemic. Lancet 398: 2126–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.CDC. Coronavirus Disease 2019 (COVID-19). Centers for Disease Control and Prevention; https://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/scientific-brief-omicron-variant.html (2020). [Google Scholar]
- 3.Investigation of SARS-CoV-2 variants: technical briefings. GOV.UK https://www.gov.uk/government/publications/investigation-of-sars-cov-2-variants-technical-briefings.
- 4.Statement – Update on COVID-19: Omicron is gaining ground: Protect, prevent, prepare. https://www.euro.who.int/en/media-centre/sections/statements/2021/statement-update-on-covid-19-omicron-is-gaining-ground-protect,-prevent,-prepare
- 5.CDC. COVID Data Tracker. Centers for Disease Control and Prevention; https://covid.cdc.gov/covid-data-tracker (2020). [Google Scholar]
- 6.Planas D, Saunders N, Maes P, Guivel-benhassine F, Planchais C, Buchrieser J, Bolland W, Porrot F, Staropoli I, Lemoine F, Péré H, Veyer D, Puech J, Rodary J, Baele G, Wawina T, Martí-carreras J, Cuypers L, Aymeric S, Prazuck T, Rey F, Simon-loriere E, Bruel T, Mouquet H, André E, and Schwartz O 2021. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature 602, 671–675 [DOI] [PubMed] [Google Scholar]
- 7.Wilhelm A, Widera M, Grikscheit K, Toptan T, Schenk B, and Pallas C 2021. Reduced Neutralization of SARS-CoV-2 Omicron Variant by Vaccine Sera and monoclonal antibodies. medRxiv 2021.12.07.21267432 [Google Scholar]
- 8.Rössler A, Riepler L, Bante D, von Laer D, and Kimpel J 2021. SARS-CoV-2 B.1.1.529 variant (Omicron) evades neutralization by sera from vaccinated and convalescent individuals. medRxiv 2021.12.08.21267491. [Google Scholar]
- 9.Maslo C, Friedland R, Toubkin M, Laubscher A, Akaloo T, and Kama B 2021. Characteristics and Outcomes of Hospitalized Patients in South Africa During the COVID-19 Omicron Wave Compared With Previous Waves. JAMA 327(6):583–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abdullah F, Myers J, Basu D, Tintinger G, Ueckermann V, Mathebula M, Ramlall R, Spoor S, De Villiers T, Van Der Walt Z, Cloete J, Rheeder P, Paruk F, Engelbrecht A, Lalloo V, Myburg M, Kistan J, Boswell MT, Gray G, Welch R, Blumberg L, and Jassat W 2021. Decreased severity of disease during the first global omicron variant covid-19 outbreak in a large hospital in tshwane, south africa. Int J Infect Dis 116:38–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McMahan K, Yu J, Mercado NB, Loos C, Tostanoski LH, Chandrashekar A, Liu J, Peter L, Atyeo C, Zhu A, Bondzie EA, Dagotto G, Gebre MS, Jacob-Dolan C, Li Z, Nampanya F, Patel S, Pessaint L, Van Ry A, Blade K, Yalley-Ogunro J, Cabus M, Brown R, Cook A, Teow E, Andersen H, Lewis MG, Lauffenburger DA, Alter G, and Barouch DH 2021. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature 590: 630–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rydyznski Moderbacher C, Ramirez SI, Dan JM, Grifoni A, Hastie KM, Weiskopf D, Belanger S, Abbott RK, Kim C, Choi J, Kato Y, Crotty EG, Kim C, Rawlings SA, Mateus J, Tse LPV, Frazier A, Baric R, Peters B, Greenbaum J, Ollmann Saphire E, Smith DM, Sette A, and Crotty S 2020. Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity. Cell 183: 996–1012.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liao M, Liu Y, Yuan J, Wen Y, Xu G, Zhao J, Cheng L, Li J, Wang X, Wang F, Liu L, Amit I, Zhang S, and Zhang Z 2020. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 26: 842–844. [DOI] [PubMed] [Google Scholar]
- 14.Jergovič M, Uhrlaub JL, Watanabe M, Bradshaw CM, White LM, LaFleur BJ, Edwards T, Sprissler R, Worobey M, Bhattacharya D, and Nikolich-Žugich J 2021. Competent immune responses to SARS-CoV-2 variants in older adults following mRNA vaccination. bioRxiv 2021.07.22.453287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tarke A, Sidney J, Methot N, Crotty S, Grifoni A, Tarke A, Sidney J, Methot N, Yu ED, Zhang Y, Dan JM, Goodwin B, Antunes S, Peters B, Scheuermann RH, Weiskopf D, and Crotty S 2021. Article Impact of SARS-CoV-2 variants on the total CD4 + and CD8 + T cell reactivity in infected or vaccinated individuals ll ll Impact of SARS-CoV-2 variants on the total CD4 + and CD8 + T cell reactivity in infected or vaccinated individuals. Cell Reports Med 2(7):100355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lutrick K, Ellingson KD, Baccam Z, Rivers P, Beitel S, Parker J, Hollister J, Sun X, Gerald JK, Komatsu K, Kim E, Lafleur B, Grant L, Yoo YM, Kumar A, Lamberte JM, Cowling BJ, Cobey S, and Thornburg NJ 2021. COVID-19 Infection, Reinfection, and Vaccine Effectiveness in Arizona Frontline and Essential Workers : Protocol for a Longitudinal Cohort Study JMIR Res Protoc. 10(6):e28925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tarke A, Coelho CH, Zhang Z, Crotty S, Grifoni A, Tarke A, Coelho CH, Zhang Z, Dan JM, Yu ED, Methot N, Weiskopf D, Antunes S, Crotty S, Grifoni A, and Sette A 2022. Article SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron ll ll Article SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell 185: 847–859.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naleway AL, Wesley MG, Gaglani M, Grant L, Caban-martinez AJ, Burgess JL, Groover K, Yoon SK, Tyner HL, Schaefer-solle N, Olsho LEW, Gerald JK, Rose S, Thiese MS, Mak J, Louzado P, Lundgren J, Groom HC, Edwards LJ, Lutrick K, Phillips AL, Mayo J, Roger L, Dunnigan K, Smith M, Sokol BE, Odean M, Ellingson KD, Dickerson M, Hegmann KT, Respet K, Fleary DE, Murthy K, Hunt A, Cruz A, Damena EA, Harder GJA, Jennifer LO, Melissa V, John A, Mistry P, Thompson MG, and Fowlkes AL 2022. Incidence of SARS-CoV-2 infection among COVID-19 vaccinated and unvaccinated healthcare personnel, first responders, and other essential and frontline workers : Eight US locations, January – September 2021. Influenza Other Respir Viruses. 2022 Jan 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grifoni A, Weiskopf D, Ramirez SI, Smith DM, Crotty S, Sette A, Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Jadi RS, Marrama D, De Silva AM, Frazier A, Carlin AF, and Greenbaum JA 2020. Article Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals ll Article Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 181: 1489–1501.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guerrera G, Picozza M, Orso SD, Placido R, Pirronello M, Verdiani A, Termine A, Fabrizio C, Giannessi F, Sambucci M, Balice MP, Caltagirone C, Salvia A, Rossini A, Battistini L, and Borsellino G 2021. BNT162b2 vaccination induces durable SARS-CoV-2 – specific T cells with a stem cell memory phenotype. Sci Immunol (2022) 6:eabl5344. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and materials that support the findings of this study are available from the corresponding author upon request.