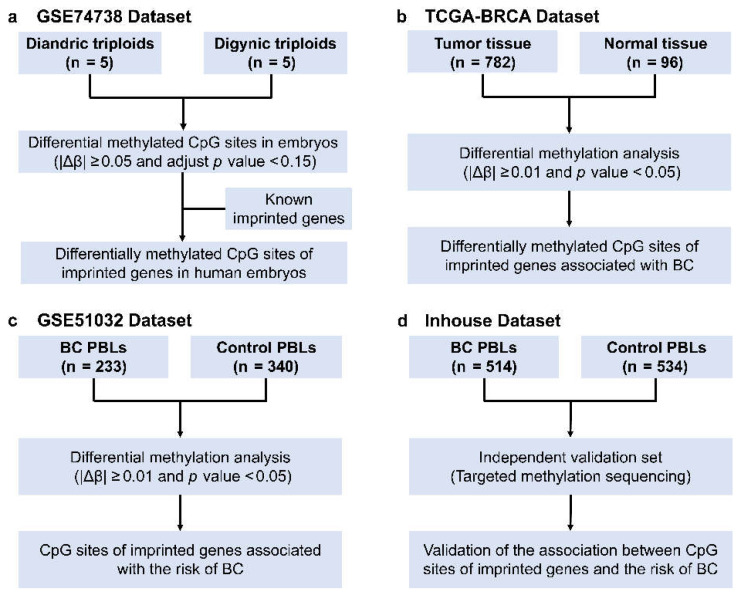
Figure 1.
Overall workflow for identification and validation in different datasets. BC, breast cancer; PBLs, peripheral blood leukocytes, TCGA, The Cancer Genome Atlas. (a) Differential methylation analyses between diandric and digynic triploids. (b) Differential methylation analyses in selected CpG sites between 782 BC tissues and 96 normal tissues in the TCGA dataset. (c) Differential methylation analyses in selected CpG sites between 233 BC patients and 340 healthy controls in the GSE51032 dataset. (d) Validation of the association between CpG sites of imprinted genes and the risk of BC in an independent case-control study.