Abstract
Background
Leiomyosarcoma is a malignant mesenchymal tumor of cells of smooth muscle lineage arising commonly in retroperitoneum, uterus, large veins, and the limbs. The genetics of leiomyosarcomas are complex and there is very limited understanding of common driver mutations. Circulating tumor DNA (ctDNA) offers a rapid and noninvasive method of next-generation sequencing (NGS) that could be used for diagnosis, therapy, and detection of recurrence.
Methods
ctDNA testing was performed using Guardant360, which detects single nucleotide variants, amplifications, fusions, and specific insertion/deletion mutations in 73 genes using NGS.
Results
Of 73 patients, 59 were found to have one or more cancer-associated genomic alteration. Forty-five (76%) were female with a median age of 63 (range, 38–87) years. All samples were designated metastatic. The most common alterations were detected in Tp53 (65%), BRAF (13%), CCNE (13%), EGFR (12%), PIK3CA (12%), FGFR1 (10%), RB1(10%), KIT (8%), and PDGFRA (8%). Some of the other alterations included RAF1, ERBB2, MET, PTEN TERT, APC, and NOTCH1. Potentially targetable mutations, by Food and Drug Administration–approved or clinical trials, were found in 24 (40%) of the 73 patients. Four patients (5%) were found to have incidental germline TP53 mutations.
Conclusion
NGS of ctDNA allows identification of genomic alterations in plasma from patients with leiomyosarcoma. Unfortunately, there is limited activity of current targeted agents in leiomyosarcomas. These results suggest opportunities to develop therapy against TP53, cell cycle, and kinase signaling pathways. Further validation and prospective evaluation is warranted to investigate the clinical utility of ctDNA for patients with leiomyosarcoma.
Keywords: ctDNA, genetic alterations, leiomyosarcoma
INTRODUCTION
Leiomyosarcoma (LMS) is a malignant mesenchymal tumor with smooth muscle cell differentiation. Because these cells are present in all organs, LMS can arise anywhere in the body, commonly in the retroperitoneum, uterus, large veins, and the limbs. It accounts for 10% to 20% of all sarcomas and has been classically reported as the most frequent soft tissue sarcoma subtype together with liposarcoma.[1,2] Uterine LMSs are the single largest site-specific group with an incidence of 0.64 per 100,000 women.[3]
The overall incidence of LMSs increases with age and peaks at the seventh decade of life. The presentation differs from uterine LMS, which occurs in women in the perimenopausal age group.[3] The differences among the gender incidence depend on the primary tumor site; retroperitoneal and inferior vena cava sites predominate in women,[4] whereas noncutaneous soft tissue sites and cutaneous LMS are more common in men.[1,5] There are no specific diagnostic clinical features of soft tissue LMS to distinguish from other soft tissue sarcomas. On suspicion, the diagnostic and staging studies are performed simultaneously with a biopsy to establish a specific diagnosis.[6] The treatment and prognosis depend on factors such as histological grade, tumor size, and location, as well as the presence of localized or metastatic disease. LMS outcomes are mostly dependent on feasibility for surgical resection at an early stage with wide margins. Once metastatic, systemic chemotherapy such as doxorubicin ± ifosfamide or dacarbazine or gemcitabine/docetaxel is used. The addition of chemotherapy offers a median progression-free survival (PFS) of approximately 6 months and an overall survival (OS) of 10 to 15 months, representing an unmet need of further therapeutic agents.[1] Pazopanib is a multikinase inhibitor approved for the second-line treatment in LMS; its use has resulted in increased PFS but no OS benefit.[7]
In the era of next-generation sequencing (NGS), there is a paucity of available targets with therapeutic potential in LMS. The exact pathophysiologic genetic framework remains elusive because of the rarity of the disease, limited NGS testing, and lack of universal standards. Various studies have been published showing complex heterogeneity with genomic instability associated with defects in TP53 and ATM gene.[1]
Circulating tumor DNA (ctDNA) offers a novel, rapid, and noninvasive method of NGS that could be used for diagnosis, therapy, and detection of recurrence. Similar studies have been published in other soft tissue sarcomas, such as gastrointestinal stromal tumors (GIST).[8] ctDNA consists of small fragments of DNA, comprising fewer than 200 nucleotides found in the bloodstream, and is a marker of cancer cell turnover. There are no oncogenic single nucleotide variants (SNVs) that characterize LMS. Still, loss of tumor suppressors, including TP53, RB1, and PTEN, are commonly seen, as are multiple copy number alterations (CNAs). Most ctDNA assays have been developed to detect SNVs that are highly recurrent in many types of carcinomas. The lack of recurrent SNVs in LMS poses a limitation for targeted sequencing; however, the numerous CNAs characteristic of this disease represent an ideal target for detection.[9,10] To date, this study is the largest evaluation of the genetic landscape for looking at LMS using ctDNA.
PATIENTS AND METHODS
This is a retrospective observational study of 73 de-identified patients from the Guardant Health Data base. Patients with diagnosis of LMS were referred from academic as well as community institutions. Patients had blood samples collected between December 2014 and December 2018 and sent for analysis to the Guardant Health Group.
The Guardant 360 is a commercially available NGS panel for identifying SNVs in 73 genes, CNAs in 18 genes, and fusions, deletions and insertions in 23 genes.
ctDNA Isolation
The 10-mL blood samples were collected in Streck tubes. The samples were then stored and shipped at room temperature to Guardant Health (Redwood City, CA, USA). The plasma was isolated by centrifugation of blood by 1600g for 10 minutes at 4°C. Using the QIAamp circulating nucleic acid kit (Qiagen, Hilden, Germany), ctDNA was then extracted, concentrated, and size selected using Agencourt Ampure XP beads (Beckman Coulter, Brea, CA, USA), and quantified by Qubit fluorometer (Life Technologies, Carlsbad, CA, USA).
ctDNA Sequencing
After isolation and oligo-nucleotide barcoding, ctDNA digital sequencing library preparation was performed. The library was then amplified and enriched for target genes. Each base pair had a 15,000x average coverage depth. After sequencing, algorithmic reconstruction of the digitized sequencing signals were used to reconstruct the ctDNA fragments.[11]
The patient demographics and tumor characteristics were obtained and subsequently stored in a secure database for analysis.
Statistical Analysis
The patient and tumor characteristics were analyzed by using simple descriptive statistics. The results of the genetic alterations were reported in the form of a bar graph.
RESULTS
A total of 73 patients were detected as having the diagnosis of LMS from the formalin-fixed tissue samples. Most of the patient cohort was composed of a female population (76%) with a mean age of 63 years (range 38–87 years). All patients were found to have metastatic leiomyosarcoma.
The most common mutation found was the TP53, present in 48 patients (65%), whereas KIT and PDGFRA were the least frequent, each present in only 5 patients (8%). In-between were BRAF and CCNE (present in 8 patients, corresponding to 13%), EGFR and PIK3CA (in 7 patients each, 12%), and FGFR1 and RB1 (6 patients, 10% of the sample). There were 14 patients with variations of undetermined significance (VUS) (Figure 1).
Figure 1.
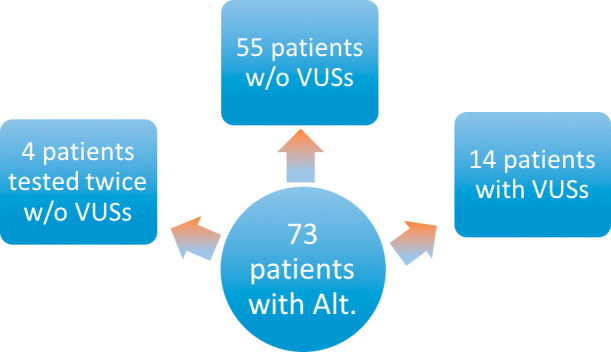
Consort flow diagram showing the number of patients with alterations. VUS, variations of undetermined significance; Alt, alterations.
The other alterations included RAF1, ERBB2, MET, PTEN, TERT, APC, and NOTCH1. Twenty-four patients (40%) had genomic alterations, detected by ctDNA, potentially targetable by a Food and Drug Administration (FDA)-approved or clinical trial therapy according to the European Society for Medical Oncology (ESMO) Scale of Clinical Actionability for molecular targets (ESCAT) and OncoKB (Table 2). There were 4 (5%) patients who were found to have incidental germline TP53 mutations. The various mutations are shown in the form a bar chart in Figure 2.
Table 2.
The targetable mutations that are approved by the Food and Drug Administration or used in the clinical trial European Society for Medical Oncology Scale of Clinical Accountability for molecular targets (ESCAT) and OncoKB
|
Mutation
|
ESCAT/OncoKB, Tier
|
| TP53 | X |
| BRAF | III |
| CCNE | X |
| EGFR | III |
| PIK3CA | III |
| FGFR1 | III |
| RB1 | X |
| KIT | III |
| PDGFRA | III |
| RAF1 | III |
| ERBB2 | III |
| MET | III |
Figure 2.
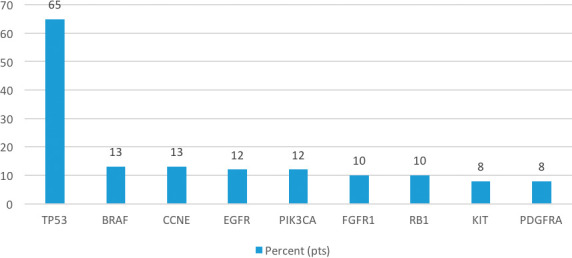
Most commonly mutated genes by ctDNA analysis of patients with metastatic leiomyosarcoma (N=73).
Table 1.
The percentage of patients with mutations and nature of mutations
|
Mutation
|
Percent (Patients)
|
Copy Number Variation Count
|
Insertion/Deletion Count
|
Single Nucleotide Variants Count
|
| TP53 | 65 (48) | - | 14 | 43 |
| BRAF | 13 (8) | 5 | - | 4 |
| CCNE | 13 (8) | 8 | - | - |
| EGFR | 12 (7) | 4 | - | 4 |
| PIK3CA Level 1 | 12 (7) | 6 | - | 1 |
| FGFR1 Level 4 | 10 (6) | 3 | - | 3 |
| RB1 | 10 (6) | - | 5 | 1 |
| KIT | 8 (5) | 5 | - | - |
| PDGFRA | 8 (5) | 4 | - | 1 |
-, not detected.
There were a total of 90 samples. Of all the samples collected and analyzed, alterations were detected in 73 ctDNA samples (>80%). After exclusion of VUS, 63 ctDNA samples harbored cancer-associated genomic alterations (70%), and of these, 59 were found to have 1 or more cancer-associated genomic alterations.
The copy number variation count (the variation between genomes in the number of copies of a genomic region) was also described, being the highest among the CCNE mutation. There were 14 insertions/deletions among the Tp53 mutation and a total of 43 SNVs on this same mutation.
DISCUSSION
LMSs are highly aggressive neoplasms with dismal prognosis in the metastatic setting: PFS of only 6 months and an OS of approximately 12 to 15 months.[12] Regimens associated with survival benefit include doxorubicin/ifosfamide[13] and gemcitabine/docetaxel.[14] The prognosis is even worse with second- and third-line chemotherapy options. This accounts for the need of new therapeutic options.
NGS offers detection of genetic alterations not only responsible for diagnosis or the pathophysiology but also provide information about therapeutic targets. The deletions in canonical cancer genes are central to LMS along with DNA repair enzymes.[15] ctDNA is a novel and rapid method of NGS, especially useful when tissue biopsy is not available or cannot be obtained. The quantity of ctDNA varies among individuals and depends on the type of tumor, its location and stage, and the disease burden. The sensitivity of ctDNA is higher in metastatic patients with high disease burden.[11] ctDNA detection is useful in diagnosis, response assessment, and disease progression.[16,17]
Presented here is the largest study identifying the genetic makeup of patients with metastatic LMS with the use of ctDNA. Meanwhile, there have been a few studies looking at the genetic framework from tissue-based NGS. Lee et al[15] described the most frequent alterations found in a series of 25 uterine and nonuterine LMS. Frequently involved genes were Tp53, ATM, ATRX, EGFR, and RB1, with the CNAs identified in 85% of cases. Another study with whole genomic sequencing and transcriptomic analysis showed that most of the alterations were mutations in tumor suppressor genes including Tp53, RB1, ATRX, and ATM; chemokine receptors including FGFR2 and ALK; chromatin modifiers, such as DNMT3A; and transcription regulators, such as PAX3. Most of genes were associated with the copy number amplification.[18] Cuppens et al[7] reported common alterations in Tp53, RB1, and PTEN in uterine LMSs.
Hemming et al[10] described the use of ctDNA in patients with progressive LMS. Here similarly, the most common alterations were deletions in Tp53 and Rb1. The study also reported the correlation between higher levels of ctDNA with tumor size and disease progression in patients with LMS using 11 patients, 11 of 16 with active LMS had detectable ctDNA, and their results suggest that ctDNA may be useful as a biomarker for a subset of uterine and extrauterine LMS. In our study, we were able to evaluate the ctDNA of 73 patients with the diagnosis of metastatic LMS. As previously discussed, use of ctDNA detection for LMS is challenging because of the lack of characteristic oncogenic SNVs. Most ctDNA assays were developed to detect highly recurrent SNVs for many types of carcinomas, and certain sarcomas.[19,20]
We analyzed samples from 73 patients; 14 were found to have VUS resulting in a mutation detection rate of 75%. The results of our study were consistent with the prior studies showing frequent mutations in tumor-suppressing genes including Tp53, BRAF, and Rb; SNV was the most common alteration followed by the copy number variation (CNV). There have been multiple prior studies highlighting the importance of CNV in the biology of LMSs. Despite the genetic heterogeneity, there are some consistent DNA copy number changes detected by comparative genetic hybridization and molecular studies. Common changes include a high frequency of losses in DNA copy numbers in 10q and 13q, gains in 17p, and the presence of tumor size–related alterations. It is believed that changes in 10q, 13q, and 17p lead to mutations in tumor suppressor genes, which may be early changes in tumorigenesis. Studies have shown that 10q23 encodes for PTEN, 10q24–25 for MX11 tumor suppressor genes. The MX11 gene negatively regulates c-myc oncogene contributing to tumor development. Changes in 13q encode genes such as RB1 at 13q14 and BRCA2 at 13q12, which are both tumor suppressor genes. Similar gains in 17p are associated with Tp53-related alterations.[21,22] ctDNA-detected genetic alterations in our study are consistent with the prior studies.
Most of the alterations in the LMS do not have a designated target available. This has been the ongoing subject matter for most clinical trials. Tp53 is the most commonly altered gene in LMS and an attractive pharmacologic target; however, there has been no successful mutant Tp53 drug despite decade-long research. There have been new advances, such as the development of Tp53 reactivating molecules including small peptide mutant Tp53 conformation stabilizers (pCAPs), CTM, PEITC, ReACp53, and ZMC1. Although these molecules have a different mechanism of action, they all target a specific subset of Tp53 mutations, restore the wild-type conformation and function of the protein, and cause tumor regression. These molecules require further studies to determine the clinical benefit.[23] A pilot trial is under way to determine the use of atorvastatin in Tp53 mutant and wild-type malignancies. Similarly, other detected genes, such as BRAF, EGFR, and PIK3CA, have available targeted agents approved for other cancer types but have no clinical benefit in LMSs. This can be explained with the concept of driver genes, mutation types, and frequency and type of cancers. Multiple clinical trials are under way to discover a therapeutic target.
In other tumors, such as GIST, mutations detected on ctDNA analysis, such as KIT/PDGFRA, have been found relevant for diagnostic purposes as well as to predict treatment responses.[11] Meanwhile, there are no available treatment targets in LMS; ctDNA in LMS can be restricted to diagnosis related to the size cutoff 5 cm irrespective of the disease progression, making it a technical sensitivity threshold. This ctDNA detection may help distinguish between leiomyoma and LMS. Similarly, positive ctDNA results after completion of the adjuvant treatment may help with detection of recurrence.[10] The detection of SNV has proven to be beneficial in detecting resistant mutations and providing important prognostic information. A highly specific LMS-specific assay Cancer Personalized Profiling by deep Sequencing (CAPP-Seq) has been designed for the SNVs, indels, and CNAs. A similar publication described that ultra-low pass whole genome sequencing assay based ctDNA detection may have prognostic significance.[24] Although different tests have different sensitivities, the test used in this publication appears to be helpful for qualitative and quantitative ctDNA analysis, especially useful in the setting of disease surveillance.[25]
The retrospective design, lack of control arm, unavailability of tumor characteristics, lack of sensitivity, and concordance data between ctDNA and the formalin-fixed paraffin-embedded tissue are some of the limitations of the study. Meanwhile, there have been advances in the improvement of sensitivity of the assay; the use of ctDNA is being prospectively studied in clinical trials in LMSs as well as other sarcomas. A combination strategy approach for targeted agents as well as the discovery of newer targets is also under way.
CONCLUSION
In conclusion, NGS of ctDNA allows identification of genomic alterations in plasma from patients with LMS. Unfortunately, to this date there is limited activity of targeted agents in LMS. Nevertheless, our results suggest opportunities to develop therapy targeting TP53, cell cycle, and kinase signaling pathways. On the other hand, ctDNA in LMS has many potential clinical uses regarding diagnosis, response to treatment, and evaluation of the risk of recurrence. Further validation and prospective evaluation are warranted to investigate the clinical utility of ctDNA for patients with LMS.
Funding Statement
Source of Support: None.
Footnotes
Gina D'Amato serves on the advisory board for Blue print, Epizyme, and Bayer. Jonathan C. Trent serves on the advisory board for Blueprint, Deciphera, Epizyme, and Daiichi. The other authors have nothing to disclose.
Based on a presentation at the American Society of Clinical Oncology 2019 Annual Meeting.
References
- 1.Duffaud F, Ray-Coquard I, Salas S, Pautier P. Recent advances in understanding and managing leiomyosarcomas. F1000prime reports . 2015;7:55. doi: 10.12703/P7-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sephien A, Mousa MS, Bui MM, Kedar R, Thomas K. Leiomyosarcoma of the inferior vena cava with hepatic and pulmonary metastases: case report. J Radiol Case Rep . 2019;13:30–40. doi: 10.3941/jrcr.v13i5.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Setia A, Kanotra S, Aggarwal R, Bhavthankar DP. Epithelioid leiomyosarcoma of uterus. BMJ Case Rep . 2012. 2012. [DOI] [PMC free article] [PubMed]
- 4.Hashimoto H, Tsuneyoshi M, Enjoji M. Malignant smooth muscle tumors of the retroperitoneum and mesentery: a clinicopathologic analysis of 44 cases. J Surg Oncol . 1985;28:177–186. doi: 10.1002/jso.2930280307. [DOI] [PubMed] [Google Scholar]
- 5.Fields JP, Helwig EB. Leiomyosarcoma of the skin and subcutaneous tissue. Cancer . 1981;47:156–169. doi: 10.1002/1097-0142(19810101)47:1<156::aid-cncr2820470127>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 6.Casali PG, Blay JY, Bertuzzi A, et al. Soft tissue and visceral sarcomas: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol . 2014;25:102–112. doi: 10.1093/annonc/mdu254. [DOI] [PubMed] [Google Scholar]
- 7.Cuppens T, Moisse M, Depreeuw J, et al. Integrated genome analysis of uterine leiomyosarcoma to identify novel driver genes and targetable pathways. Int J Cancer . 2018;142:1230–1243. doi: 10.1002/ijc.31129. [DOI] [PubMed] [Google Scholar]
- 8.Arshad J, Roberts A, Nagy RJ, Wilky BA, Trent JC. Utility of circulating tumor DNA (ctDNA) in the management of patients with gastrointestinal stromal tumor (GIST): Analysis of 152 patients. J Clin Oncol . 2018;36(15_suppl):11539–11539. [Google Scholar]
- 9.Wan JCM, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer . 2017;17:223–238. doi: 10.1038/nrc.2017.7. [DOI] [PubMed] [Google Scholar]
- 10.Hemming ML, Klega KS, Rhoades J, et al. Detection of circulating tumor DNA in patients with leiomyosarcoma with progressive disease. JCO Precis Oncol . 2019. 2019. [DOI] [PMC free article] [PubMed]
- 11.Arshad J, Roberts A, Ahmed J, et al. Utility of circulating tumor DNA in the management of patients with GI stromal tumor: analysis of 243 patients. JCO Precis Oncol . 2020. pp. 66–73. [DOI] [PubMed]
- 12.Penel N, Italiano A, Isambert N, Bompas E, Bousquet G, Duffaud F. Factors affecting the outcome of patients with metastatic leiomyosarcoma treated with doxorubicin-containing chemotherapy. Ann Oncol . 2010;21:1361–1365. doi: 10.1093/annonc/mdp485. [DOI] [PubMed] [Google Scholar]
- 13.Sleijfer S, Ouali M, van Glabbeke M, et al. Prognostic and predictive factors for outcome to first-line ifosfamide-containing chemotherapy for adult patients with advanced soft tissue sarcomas: an exploratory, retrospective analysis on large series from the European Organization for Research and Treatment of Cancer-Soft Tissue and Bone Sarcoma Group (EORTC-STBSG) Eur J Cancer . 2010;46:72–83. doi: 10.1016/j.ejca.2009.09.022. [DOI] [PubMed] [Google Scholar]
- 14.Hensley ML, Maki R, Venkatraman E, et al. Gemcitabine and docetaxel in patients with unresectable leiomyosarcoma: results of a phase II trial. J Clin Oncol . 2002;20:2824–2831. doi: 10.1200/JCO.2002.11.050. [DOI] [PubMed] [Google Scholar]
- 15.Lee PJ, Yoo NS, Hagemann IS, et al. Spectrum of mutations in leiomyosarcomas identified by clinical targeted next-generation sequencing. Exp Mol Pathol . 2017;102:156–161. doi: 10.1016/j.yexmp.2017.01.012. [DOI] [PubMed] [Google Scholar]
- 16.Batth IS, Mitra A, Manier S, et al. Circulating tumor markers: harmonizing the yin and yang of CTCs and ctDNA for precision medicine. Ann Oncol . 2017;28:468–477. doi: 10.1093/annonc/mdw619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arshad JA, Subhawong T, Trent JC. Progress in determining response to treatment in gastrointestinal stromal tumor. Expert Rev Anticancer Ther . 2020;4:279–288. doi: 10.1080/14737140.2020.1745068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chudasama P, Mughal SS, Sanders MA, et al. Integrative genomic and transcriptomic analysis of leiomyosarcoma. Nat Commun . 2018;9:144. doi: 10.1038/s41467-017-02602-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Azad TD, Chaudhuri AA, Fang P, et al. Circulating tumor DNA analysis for detection of minimal residual disease after chemoradiotherapy for localized esophageal cancer. Gastroenterology . 2019;158:494–505. doi: 10.1053/j.gastro.2019.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.An Y, Guan Y, Xu Y, et al. The diagnostic and prognostic usage of circulating tumor DNA in operable hepatocellular carcinoma. Am J Transl Res . 2019;11:6462–6474. [PMC free article] [PubMed] [Google Scholar]
- 21.El-Rifai W, Sarlomo-Rikala M, Knuutila S, Miettinen M. DNA copy number changes in development and progression in leiomyosarcomas of soft tissues. Am J Pathol . 1998;153:985–990. doi: 10.1016/S0002-9440(10)65640-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Svarvar C, Larramendy ML, Blomqvist C, et al. Do DNA copy number changes differentiate uterine from non-uterine leiomyosarcomas and predict metastasis? Mod Pathol . 2006;19:1068–1082. doi: 10.1038/modpathol.3800617. [DOI] [PubMed] [Google Scholar]
- 23.Kogan S, Carpizo D. Pharmacological targeting of mutant p53. Transl Cancer Res . 2016;5:698–706. doi: 10.21037/tcr.2016.11.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shulman DS, Klega K, Imamovic-Tuco A, et al. Detection of circulating tumour DNA is associated with inferior outcomes in Ewing sarcoma and osteosarcoma: a report from the Children's Oncology Group. Br J Cancer . 2018;119:615–621. doi: 10.1038/s41416-018-0212-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Przybyl J, Chabon JJ, Spans L, et al. Combination approach for detecting different types of alterations in circulating tumor DNA in leiomyosarcoma. Clin Cancer Res . 2018;24:2688–2699. doi: 10.1158/1078-0432.CCR-17-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]


