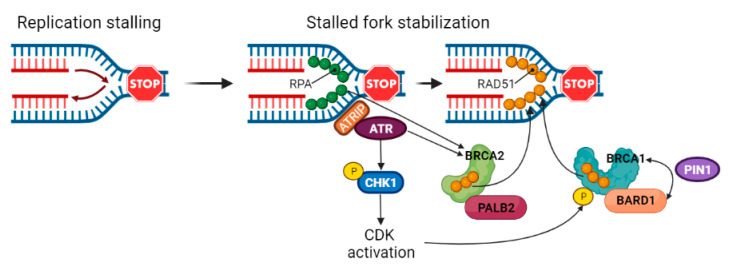
Figure 4.
Stalled fork stabilization. During DNA duplication, replication forks often encounter obstacles that can lead to genotoxic fork stalling. Stalled forks are characterized by exposed DNA ends, which can be degraded by various cellular nucleases. Under replication stress, extensive ssDNA is readily protected and stabilised by RPA. ATRIP binding to RPA–ssDNA promotes ATR localization. ATR phosphorylates RPA and CHK1, leading to CDK activation. Both ATR and RPA promote PALB2 and BRCA2 translocation to stalled replication forks, while CDK phosphorylates BRCA1. The RPA–ssDNA complex is displaced by the RAD51 protein. RAD51 loading and stabilization are enhanced by the BRCA2/PALB2 complex as well as by the BRCA1–BARD1 complex. The phosphorylation-directed prolyl isomerase PIN1 enhances BRCA1–BARD1 interaction with RAD51. Stable RAD51 nucleoprotein filaments then mediate replication fork reversal.