Abstract
Simple Summary
The canonical mutations in gastrointestinal stromal tumors (GISTs) are typically activating mutations in KIT and platelet-derived growth factor receptor alpha (PDGFRA). Imatinib, the treatment of choice for GISTs, shows a lower response in KIT/PDGFRA wild-type GISTs. Neurotrophic tyrosine receptor kinase (NTRK) fusion, which can be treated with an NTRK target agent, has been reported in KIT/PDGFRA wild-type GISTs, and, therefore, the Yonsei Cancer Center analyzed NTRK fusion incidence in KIT/PDGFRA wild-type GISTs. At the Yonsei Cancer Center, NTRK fusion was confirmed in 16% of cases. Confirmation of NTRK fusion in KIT/PDGFRA wild-type GISTs provides important information for improving therapeutic outcomes. NTRK fusion was confirmed in 16% of KIT/PDGFRA wild-type GIST cases at the Yonsei Cancer Center. Confirmation of NTRK fusion in KIT/PDGFRA wild-type GISTs will improve therapeutic outcomes.
Abstract
The canonical mutations in gastrointestinal stromal tumors (GISTs) are typically activating mutations in KIT and platelet-derived growth factor receptor alpha (PDGFRA). GISTs with non-canonical mutations are a heterogeneous group. Here, we examined tropomyosin-related kinase (TRK) fusion in GIST cases without KIT/PDGFRA mutations (KIT/PDGFRA wild-type (WT) GISTs). We retrospectively analyzed patients who were diagnosed with GISTs at the Yonsei Cancer Center, Severance Hospital, between January 1998 and December 2016. Thirty-one patients with KIT/PDGFRA WT GISTs were included in the analysis. TRK expression in tumor samples was assessed by pan-TRK immunohistochemistry (IHC), and the neurotrophic tyrosine receptor kinase (NTRK: the gene encoding TRK) rearrangement was analyzed by fluorescence in situ hybridization (FISH). IHC analyses revealed that five cases in this cohort exhibited a weak to moderate TRK expression. NTRK1 fusions were detected in three tumor samples, and two samples harbored NTRK3 fusions. The remaining 26 samples did not harbor NTRK fusions. Two types of NTRK fusions were detected, and the overall NTRK fusion frequency in KIT/PDGFRA WT GIST cases was 16% (5/31). Our data provide insights into the molecular alterations underpinning KIT/PDGFRA WT GISTs. More effort should be devoted to improve methods to identify this distinct disease subtype within the KIT/PDGFRA WT GIST group.
Keywords: gastrointestinal stromal tumor, KIT, PDGF receptor tyrosine kinase, neurotrophic tyrosine receptor kinase fusion, tropomyosin-related kinase fusion
1. Introduction
Gastrointestinal stromal tumors (GISTs) are the most common mesenchymal tumors of the GI tract, arising from the interstitial Cajal cells [1]. These tumors account for approximately 20% of soft tissue sarcomas, and have an annual incidence of approximately 10 per 1 million individuals [2,3]. Activating mutations in KIT and platelet-derived growth factor receptor alpha (PDGFRA) are considered to be the main oncogenic drivers of GISTs; most GISTs (75%) harbor a mutation in KIT, and PDGFRA mutations occur in 10% to 20% of GISTs [2]. Cases lacking these mutually exclusive mutations are less common, and have been classified as KIT/PDGFRA wild-type (WT) GISTs [4]. KIT/PDGFRA WT GISTs are a heterogeneous group of different diseases composed of various clinical phenotypes and molecular characteristics [5].
Imatinib mesylate and other small-molecule tyrosine kinase inhibitors (TKIs), such as sunitinib and regorafenib, have demonstrated anticancer activity in GISTs. Therefore, these drugs have been established as part of the protocol for the treatment of GIST patients. Imatinib is the only TKI approved for both the adjuvant and palliative treatment of GISTs, and it is the first-line therapy for unresectable GISTs. Sunitinib is the second-line therapy for patients with imatinib resistance or intolerance, and regorafenib is the third-line therapy for patients with unresectable metastatic GISTs that no longer respond to imatinib and sunitinib [3]. However, patients with advanced GISTs have different responses to imatinib. Advanced WT GISTs have a 0–45% likelihood of responding to imatinib [6]. Additionally, patients with KIT/PDGFRA WT GISTs are unlikely to benefit from imatinib treatment [3].
Tropomyosin-related kinase (TRK) A, TRKB, and TRKC are receptor tyrosine kinases encoded by neurotrophic receptor tyrosine kinase (NTRK) 1, NTRK2, and NTRK3, respectively. TRK fusions occur in diverse cancers in children and adults [7]. Two studies have reported that the ETS variant transcription factor 6 (ETV6)-NTRK3 fusion is an actionable target in KIT/PDGFRA WT GISTs [8,9]. Recently, the first-generation TRK inhibitors larotrectinib and entrectinib were shown to have marked and durable antitumor activity in patients with TRK fusion-positive cancers, regardless of the age of the patient or the tumor type [10,11]. Therefore, it is important to identify which KIT/PDGFRA WT GIST patients can benefit from treatment with this type of drug. We investigated the TRK fusion status in patients with KIT WT GISTs using immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH) to determine whether we could identify patients with KIT/PDGFRA WT GISTs who may be potential candidates for TRK inhibitors.
2. Materials and Methods
2.1. Patients
We retrospectively analyzed the records of patients who were diagnosed with a GIST at the Yonsei Cancer Center, Severance Hospital, between January 1998 and December 2016. At the time of diagnosis, all cases underwent immunohistochemistry (IHC) for C-kit (CD117), CD34, smooth muscle actin (SMA), and S-100 protein. All 38 KIT and PDGFRA WT GISTs showed diffuse or focal immunoreactivity for C-kit (CD117) and CD34, while the IHC results for SMA and S-100 protein were negative. Distinct histologic features were not observed. Detailed information on patient characteristics was obtained from an electronic medical database. The observation period lasted until November 2019. This study was approved by the Severance Hospital Institutional Review Board (4-2017-0605). The requirement for written informed consent was waived, given the retrospective nature of the study.
2.2. Treatment
A total of 38 patients was found to have a KIT WT GIST, and 31 patients with sufficient intact tissue samples were included in the analysis. Twenty-eight patients underwent surgery. Eight patients received imatinib as adjuvant therapy, and two patients received neoadjuvant imatinib followed by adjuvant imatinib. Of the three patients who did not undergo surgery, two were treated with imatinib, and one refused treatment. If patients started developing resistance to imatinib, the dose was increased to up to 800 mg/day. If disease progression occurred with the maximum dose of imatinib (800 mg/day), the treatment regimen was changed, if possible.
2.3. KIT and PDGFRA Analysis
A commercial sequencing service was used for KIT and PDGFRA analyses. The lab used Sanger sequencing (direct sequencing). Exons 9, 11, 13, and 17 for KIT and exons 12 and 18 for PDGFRA were included for mutation analyses.
2.4. IHC for TRK Expression
Archival formalin-fixed paraffin-embedded (FFPE) tissues were obtained and, for each case, a block containing a representative lesion was chosen. Tissue sections from these blocks were used for IHC, which was performed using a Ventana XT automated immunohistochemical staining instrument (Ventana Medical Systems, Tucson, AZ, USA) according to the manufacturer’s protocol. Briefly, 4 μM thick sections were deparaffinized using EZ Prep solution (Ventana Medical Systems) and incubated in a CC1 standard solution (pH 8.4 buffer containing Tris/borate/EDTA) for antigen retrieval. Sections were blocked in Inhibitor D (3% H2O2) for 4 min at 37 °C to block endogenous peroxidases. Slides were incubated with the primary antibody for 24 min at 37 °C, and then with a universal secondary antibody for 8 min at 37 °C. The primary antibody used for IHC staining was anti-pan-TRK (1:100; EPR17341; rabbit monoclonal; Abcam, Cambridge, MA, USA), which detects TRKA, TRKB, and TRKC. The slides were then incubated in streptavidin–horseradish peroxidase for 4 min at 37 °C, followed by a 4 min incubation in the substrate, a solution containing 3,3′-diaminobenzidine tetrahydrochloride and H2O2. Finally, the slides were counterstained with hematoxylin and a bluing reagent at 37 °C.
The stained slides were evaluated for the proportion and intensity of pan-TRK staining. The intensity of cytoplasmic staining was scored as negative (0), weakly positive (1), moderately positive (2), or strongly positive (3). The proportion of staining was scored based on the percentage of positive cells. The final score was obtained using the formula: 3 × percentage of strongly positive cells + 2 × percentage of moderately positive cells + 1 × percentage of weakly positive cells. The final H-score was a value within the range of 0 to 300.
2.5. FISH
All GIST samples were screened using commercially available NTRK1, NTRK2, and NTRK3 split FISH probes (Abnova, Taipei, Taiwan), which detect the rearrangements of these genes. The FISH analysis was performed using 2 µM thick FFPE tissue sections. At least 100 nuclei were evaluated by an experienced pathologist for each sample. NTRK1, NTRK2, and NTRK3 with no gene rearrangements showed closely located red and green signals. If more than 15 of 100 nuclei demonstrated separately located red and green signals, this was deemed to indicate NTRK rearrangements (i.e., a cutoff value of 15%) [12].
3. Results
3.1. Patient Characteristics and Treatment
A total of 821 patients was diagnosed with GISTs between January 1998 and December 2016. Thirty-eight patients had KIT and PDGFRA WT GISTs, and 31 patients had enough intact tissue samples for analysis. Those 31 patients with KIT and PDGFRA WT GISTs were included in this study. Twenty-eight patients underwent curative resection with or without neoadjuvant and/or adjuvant imatinib, according to the physician’s decision. Three patients had unresectable disease, of whom two received palliative imatinib and one refused any further treatment. The patients’ characteristics are summarized in Table 1. The median age at diagnosis was 56 years (range: 33–92 years); 14 patients (45.2%) were men and 17 (54.8%) were women. The median follow-up duration was 50 months (range: 3–132 months). Four patients died and three patients relapsed after surgery or adjuvant therapy with imatinib. Of the three patients with recurrent disease, one received adjuvant therapy with imatinib.
Table 1.
Baseline characteristics.
| Characteristics | Total (n = 31) N (%) | NTRK Fusion (n = 5) | NTRK Wild Type (n = 26) | p-Value |
|---|---|---|---|---|
| Age | ||||
| Median (range) | 56 (33–92) | 61 (33–92) | 53 (37–84) | 0.163 |
| Sex | - | - | - | 1000 |
| Male | 14 (45.2) | 2 (40.0) | 12 (46.2) | - |
| Female | 17 (54.8) | 3 (60.0) | 14 (53.8) | - |
| Tumor size (cm) | - | - | - | - |
| Median ± SD | 8.2 ± 5.3 | 3.9 ± 6.5 | 6.0 ± 5.2 | 0.410 |
| Tumor location | - | - | - | 0.028 |
| Abdominal wall | 1 (0.3) | 0 (0.0) | 1 (3.8) | - |
| Stomach | 14 (45.1) | 1 (20.0) | 14 (53.8) | - |
| Small bowel | 11 (35.4) | 2 (40.0) | 6 (23.1) | - |
| Descending colon | 1 (0.3) | 0 (0.0) | 1 (3.8) | - |
| Rectum | 4 (12.9) | 2 (40.0) | 4 (15.4) | - |
| Tumor size, groups (cm) ◇ | - | - | - | 1000 |
| ≤5 | 14 (50.0) | 3 (60.0) | 11 (47.8) | - |
| >5 | 14 (50.0) | 2 (40.0) | 12 (52.2) | -- |
| Mitotic rate ◐ (no. of mitoses/50 HPFs *) | -- | 0.711 | ||
| ≤5 | 11 (50.0) | 2 (40.0) | 12 (52.2) | - |
| >5 | 16 (50.0) | 3 (60.0) | 10 (43.5) | - |
| Disease status | - | - | - | 0.859 |
| No evidence of disease | 21 (67.7) | 5 (100.0) | 16 (61.5) | - |
| on anticancer treatment | 3 (9.7) | 0 (0.0) | 3 (11.5) | - |
| Unknown | 3 (9.7) | 0 (0.0) | 3 (11.5) | - |
| Dead | 4 (12.9) | 0 (0.0) | 4 (15.4) | - |
| Risk of metastasis ◈ | - | - | - | 0.253 |
| Low | 8 (25.8) | 1 (20.0) | 7 (26.9) | - |
| Intermediate | 3 (9.7) | 0 (0.0) | 3 (11.5) | - |
| High | 17 (54.8) | 4 (80.0) | 13 (50.0) | - |
| Not assessable | 3 (9.7) | 0 (0.0) | 3 (11.5) | - |
| Surgical treatment | - | - | - | 1000 |
| Yes | 28 (90.3) | 5 (100.0) | 23 (88.5) | - |
| No | 3 (9.7) | 0 (0.0) | 3 (11.5) | - |
◇ For three inoperable cases, size information and mitotic rate are missing because there was no surgical specimen. ◐ Mitotic rate was not evaluated in one surgical specimen. ◈ Risk of metastasis was evaluated according to the National Institute of Health (NIH) classification. HPFs: high-power fields; * per 50 HPFs is a total of 5 mm2.
When comparing the baseline characteristics of patients with NTRK fusion and WT patients, there was a statistically significant difference between the two groups. In NTRK WT patients, the tumor location was on the stomach in 53.8% of cases, whereas in patients with NTRK fusion, 20% had the tumor in the stomach, 40% in the small bowel, and 40% in the rectum.
3.2. TRK Expression in KIT WT GISTs
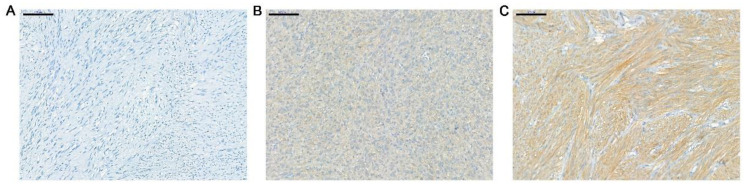
The expression of TRK in the 31 tumor samples was assessed using pan-TRK IHC. None of the samples exhibited tumor cells with a strong pan-TRK expression, and only five showed a weak to moderate pan-TRK expression (final score: 20–80) (Table 2). Representative images of TRK expression are presented in Figure 1.
Table 2.
pan-TRK IHC results.
| pan-TRK IHC | Intensity of Staining | H-Score | |||
|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | - | |
| Proportion of staining (%) | 20% | 80% | - | - | 80 |
| 20% | 80% | - | - | 80 | |
| 90% | - | 10% | - | 20 | |
| 70% | - | 30% | - | 60 | |
| 70% | - | 30% | - | 60 | |
IHC: immunohistochemistry; TRK: tropomyosin-related kinase.
Figure 1.
Representative images of pan-TRK expression in samples of WT GIST. IHC results showing samples in which the intensity of cytoplasmic staining was scored as negative (A), weakly positive (B), and moderately positive (C). There were no cases scored as strongly positive. Each scale bar is 100 μM. TRK: tropomyosin-related kinase; WT: wild type; GIST: gastrointestinal stromal tumor; IHC: immunohistochemistry.
3.3. NTRK Fusion Analyses
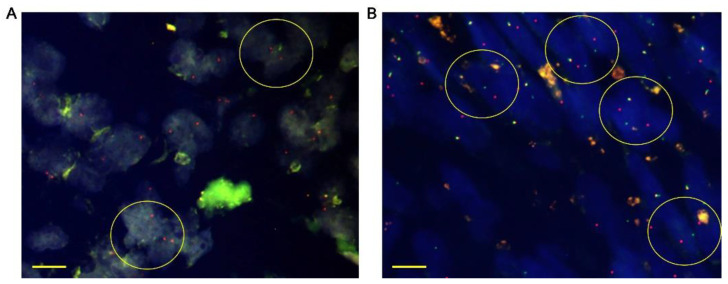
Tumor samples from 31 patients were tested for NTRK1, NTRK2, and NTRK3 fusion by FISH analysis. NTRK1 fusion was detected in three tumor samples, and two samples harbored NTRK3 fusions. The remaining 26 samples did not harbor NTRK fusions. In the five tumor samples harboring NTRK fusions, only one also tested weakly positive for TRK expression. Representative images of NTRK fusions are presented in Figure 2. The circles indicate NTRK1 fusions (Figure 2A) and NTRK3 fusions (Figure 2B).
Figure 2.
Representative images of NTRK fusion detected by FISH. Tumor tissues were stained with dual-color FISH probes. The red and green signals represent upstream and downstream probes, respectively. FISH results showing (A) NTRK1 and (B) NTRK3 fusions. Each scale bar is 10 μM. Circles indicate gene rearrangements. NTRK: neurotrophic tyrosine receptor kinase; FISH: fluorescence in situ hybridization.
3.4. Characteristics of the Five Patients Harboring NTRK Fusions
The characteristics of the five patients harboring NTRK fusions are presented in Table 3. All the patients underwent curative resection. Imatinib was not available as a therapeutic option when patient #1 underwent surgery. The disease recurred as a perineal mass 9 years after surgery, which was confirmed by biopsy. Her physician recommended imatinib as a palliative therapy, but the patient sought a second opinion. She died from disease progression 3 years after relapse, but there were no treatment details in her medical record. Patient #2 was followed up for 6 years with no evidence of disease and stopped seeing his surgeon, but he is alive and still regularly visits his physicians for other health issues at the same hospital. Patient #3 received adjuvant therapy with imatinib for 1 year after the surgery. The disease recurred 1 year and 2 months after the discontinuation of imatinib. Palliative therapy with imatinib was administered immediately after the diagnosis of recurrence and maintained for 4 years and 10 months. However, she died 2 months after the discontinuation of the drug due to her poor performance. Patient #4 received adjuvant therapy with imatinib for 3 years with no evidence of disease, and she is alive and is followed up on a regular basis. Patient #5 was followed up with for 3 years without evidence of disease in the absence of adjuvant therapy. Commercial RNA-based next-generation sequencing (NGS) (Ion Torrent Dx System, Thermo Fisher Scientific, Waltham, MA, USA) was performed using his resected tumor tissue as recommended by his physician, which revealed an ETV6-NTRK3 fusion. Images of an ETV6-NTRK3 fusion detected by FISH can be found in Figure S1.
Table 3.
Characteristics of wild-type patients harboring NTRK fusions.
| Patients (#1–#5) | #1 | #2 | #3 | #4 | #5 |
|---|---|---|---|---|---|
| Age (years) | 44 | 45 | 65 | 61 | 43 |
| Sex | F | M | F | F | M |
| Location | Rectum | Duodenum | Stomach | Jejunum | Rectum |
| Mitotic rate (no. of mitoses/50 HPFs * | 17 | 1 | 70 | 12 | 0 |
| Size (cm) | 2.8 | 1.7 | 17.0 | 3.9 | 11.0 |
| Surgery (yes or no) | yes | yes | yes | yes | yes |
| Follow-up period (years) | 11 | 6 | 8 | 4 | 3 |
| NTRK fusion by FISH | NRTK1 | NRTK3 | NRTK1 | NRTK1 | NTRK3 |
| Imatinib | No | No | Adjuvant | Adjuvant | No |
| Disease/survival status | DDP | NED | DDP | NED | NED |
NTRK: neurotrophic tyrosine receptor kinase; FISH: fluorescence in situ hybridization; HPFs: high-power fields; * per 50 HPFs is a total of 5 mm2; DDP: dead due to disease progression; NED: no evidence of disease. # Number of patient.
4. Discussion
This study reports on NTRK fusions present in patients with KIT/PDGFRA WT GISTs. In total, 16% of the WT GIST patients (5/31) had a TRK expression, and 16% (5/31) tested positive for NTRK1 (3/31: patients #1, #3, and #4) or NTRK3 (2/31: patients #2 and #5) fusions. Among the five patients harboring NTRK fusions, an ETV6-NTRK3 chimeric transcript was detected in one patient (patient #5) using commercial RNA-based NGS.
Fusions of the NTRK genes have been clinically identified and reported in various cancers [7,13]. Since TPM3-NTRK1 was first described in a human colorectal carcinoma [14], NTRK fusions have been detected in lung cancer [15], papillary thyroid carcinoma [16], colorectal cancer [17], sarcoma (including GIST) [8,9,10], and rare tumors such as secretory breast cancer [18] and mammary analog secretory carcinoma (MASC) [19]. Fusions involving NTRK1, NTRK2, or NTRK3 are the most common oncogenic alterations promoting TRK activation, and the mechanisms of NTRK gene fusion are remarkably consistent. Typically, intra- or inter-chromosomal rearrangements form hybrid genes, in which the 3′ sequences of NTRK1, NTRK2, or NTRK3, which include the kinase domain, are fused to the 5′ sequences of a different gene. This fusion produces a chimeric oncoprotein resulting in the ligand-dependent constitutive activation of TRK [20]. These fusions are thought to activate the downstream TRK kinase domain and facilitate aberrant TRK signaling via dimerization (e.g., coiled-coil domain, zinc finger domain, or WD domain). However, alternative mechanisms of dimerization or other unknown mechanisms may also exist. We attempted to perform RNA-based NGS with tumor samples obtained from all 31 patients included in this study, but most of the RNA samples did not meet the quality control criteria. We did identify ETV-NTRK3 fusion in patient #5 (unpublished data), confirming the results from previous NGS. Two different groups have previously reported that the ETV6-NTRK3 fusion is a targetable alteration in GISTs lacking mutations in the KIT/PDGFRA/RAS pathway genes (e.g., BRAF, KRAS, and NF1) and SDH deficiencies [8,9]. Brenca et al. suggested that ETV6-NTRK3 may trigger the insulin-like growth factor 1 receptor signaling cascade and the alternative nuclear insulin receptor substrate 1 pathway to promote the development of GISTs [8]. ETV6-NTRK3 fusions have also been reported in leukemia, thyroid cancer, pediatric glioma, secretory breast cancer, congenital mesoblastic nephroma, and MASC [21]. NTRK1 fusions were detected in this study, but we were unable to identify the upstream fusion partners due to the poor quality of RNA for sequencing.
Many clinical trials involving TRK inhibitors to target tumors harboring NTRK fusions are underway. A recent trial showed that LOXO-101, a pan-TRK inhibitor also called larotrectinib, exhibited marked and durable efficacy against TRK fusion-positive cancers [10], which led to fast-tracked approval by the Food and Drug Administration. A pooled analysis of three phase 1/2 clinical trials with larotrectinib suggested that TRK fusions define a unique molecular subgroup of advanced solid tumors, against which this drug is highly effective [22]. Entrectinib was also approved for patients with solid tumors harboring NTRK fusions. In this study, patient #5′s physician did not administer imatinib as an adjuvant therapy, because he had a KIT/PDGFRA WT GIST. If he experiences disease recurrence, a TRK inhibitor or other available drugs targeting NTRK fusion-positive cancers should be administered.
NTRK fusion-positive cancers can be grouped into two general categories according to the frequency at which these fusions are detected. In the first category, rare cancer types are highly enriched for NTRK fusions, including secretory breast carcinoma, MASC, congenital mesoblastic nephroma, and infantile fibrosarcoma, with a fusion rate of >90%. In the second category, NTRK fusions are found at much lower frequencies (5–25% or <5%) in more common cancers such as breast, lung, and colorectal cancers and melanoma. KIT/PDGFRA WT GISTs are also among the tumor types that have been shown to harbor NTRK fusions with frequencies of 5–25% [20]. In our study, NTRK fusions were detected in 16% of KIT WT GISTs. Therefore, despite the rarity of NTRK fusions in KIT/PDGFRA WT GISTs, it is clinically important to identify patients harboring this targetable biomarker. The accurate and efficient identification of GIST patients who are highly likely to benefit from drugs targeting NTRK fusions is critical for successful therapy.
There are different testing methods currently available to detect NTRK gene fusions in tumor samples. They include IHC, FISH, reverse transcriptase polymerase chain reaction, and NGS using DNA or RNA. Several groups have suggested workflows to detect NTRK fusions, which vary depending on the TRK expression and the incidence of NTRK fusions [23,24]. FISH is a well-established method that has been used in both clinical trials and clinical practice to test for NTRK fusions. Interestingly, Solomon et al. recently proposed a method to triage specimens based on histology and other molecular findings that efficiently identify tumors harboring these treatable oncogenic fusions. FISH, as well as RNA-level fusion testing, were suggested as confirmatory tests [24]. Penault-Llorca et al. [25] reviewed and discussed several testing methods for NTRK fusion, and proposed a testing algorithm to identify patients with TRK fusion cancers. According to the testing algorithm, FISH and NGS could be used as confirmative testing methods with pan-TRK IHC as a screening test, and pan-TRK IHC results might be helpful for selecting the confirmatory test methods, such as a targeted NGS vs. broad NGS panel. They also mentioned that tumors harboring NTRK3 rearrangements may have a weaker or false negative expression for pan-TRK IHC, and negative results from FISH or pan-TRK IHC should be confirmed by NGS. For tumors with a low incidence of NTRK fusions, such as GISTs, broad NGS testing is preferred in most suggested diagnostic algorithms [26,27,28], although FISH is also recommended as a confirmatory test. We initially planned to use IHC, FISH, and NGS to detect NTRK fusions, with the aim of using IHC as a screening method and employing FISH or NGS as confirmatory tests, depending on the circumstances of each patient. However, the RNA samples did not meet the quality control criteria, because they were retrieved from archived tissue that had been stored for years. In addition, there was discordance between the IHC and FISH results in our study.
However, other pan-TRK antibodies resolved this issue in other studies, which suggests the discordance could have been caused by the pan-TRK antibody that we used. Unfortunately, there were insufficient tumor samples to repeat the IHC.
This study had certain limitations, such as its retrospective nature, relatively small sample size, heterogeneity in tumor sampling, discordance among NTRK fusion detection methods, and selection bias, since patients were recruited from a single institution in South Korea. Furthermore, the NTRK fusion test was confirmed in GISTs, which were wild-type, not only for KIT and PDGFRA, but also for the BRAF gene [29]; however, the BRAF gene test was not performed in this study. The BRAF wild type was also not tested, which was another limitation of this study.
Nevertheless, our data provide more insights into KIT/PDGFRA WT GISTs. The TRK fusion rate of 16% is significant, because TRK is a target with effective targeted therapeutics that have been verified in many recent studies. Mesenchymal tumors driven by NTRK fusions are clinically and morphologically heterogeneous. With an increasing number of clinicopathological entities being associated with NTRK fusions, the diagnostic and predictive value of the identification of NTRK fusions is uncertain. Recently, mesenchymal tumors in the gastrointestinal tract with NTRK fusions were described as gastrointestinal stromal tumors (GISTs), but the nosology of such neoplasms remains controversial. Mesenchymal tumors of the gastrointestinal tract with NTRK rearrangements are clinically and morphologically heterogeneous, and few, if any, seem related to GISTs [30]. More studies to elucidate the genetic profile of KIT/PDGFRA WT GISTs may significantly improve the treatment outcomes of this group of GIST patients.
5. Conclusions
In conclusion, our results provided insights into the molecular alterations underpinning KIT/PDGFRA WT GISTs. Although KIT/PDGFRA WT GISTs are rare, our clinical predictions could aid physicians in identifying patients eligible for NTRK fusion screening and therapeutic targeting. More effort should be devoted to improving methods to target this distinct disease subtype within the KIT/PDGFRA WT GIST group.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/2072-6694/14/11/2659/s1, Figure S1. representative images of ETV6-NTRK3 fusion by FISH analysis of patient #5. Each scale bar is 10 μM.
Author Contributions
J.H.L. contributed to the data acquisition, writing the paper, and statistical analysis. S.-J.S. participated in patient management and data collection. E.-A.C. contributed to the data acquisition, writing the paper, and statistical analysis. J.K. participated in data collection. W.J.H. participated in patient management and data collection. H.S.K. participated in patient management and data collection. M.J. participated in patient management and data collection. S.-H.B. participated in patient management and data collection. T.I.K. participated in patient management and data collection. J.B.A. participated in patient management and data collection. H.C.C. participated in conceptualization, patient management, and data collection. S.J.S.: participated in conceptualization, patient management, and data collection. All authors reviewed the paper and approved the final version. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Severance Hospital Institutional Review Board (4-2017-0605, and date of approval 25 July 2017).
Informed Consent Statement
The requirement for written informed consent was waived, given the retrospective nature of the study.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Corless C.L. Gastrointestinal stromal tumors: What do we know now? Mod. Pathol. 2014;27((Suppl. S1)):S1–S16. doi: 10.1038/modpathol.2013.173. [DOI] [PubMed] [Google Scholar]
- 2.Ducimetiere F., Lurkin A., Ranchere-Vince D., Decouvelaere A.V., Peoch M., Istier L., Chalabreysse P., Muller C., Alberti L., Bringuier P.P., et al. Incidence of sarcoma histotypes and molecular subtypes in a prospective epidemiological study with central pathology review and molecular testing. PLoS ONE. 2011;6:e20294. doi: 10.1371/journal.pone.0020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Von Mehren M., Joensuu H. Gastrointestinal stromal tumors. J. Clin. Oncol. 2018;36:136–143. doi: 10.1200/JCO.2017.74.9705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corless C.L., Fletcher J.A., Heinrich M.C. Biology of gastrointestinal stromal tumors. J. Clin. Oncol. 2004;22:3813–3825. doi: 10.1200/JCO.2004.05.140. [DOI] [PubMed] [Google Scholar]
- 5.Brcic I., Argyropoulos A., Liegl-Atzwanger B. Update on Molecular Genetics of Gastrointestinal Stromal Tumors. Diagnostics. 2021;11:194. doi: 10.3390/diagnostics11020194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Von Mehren M., Randall R.L., Benjamin R.S., Boles S., Bui M.M., Ganjoo K.N., George S., Gonzalez R.J., Heslin M.J., Kane J.M., et al. Soft tissue sarcoma, version 2.2018, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Cancer Netw. 2018;16:536–563. doi: 10.6004/jnccn.2018.0025. [DOI] [PubMed] [Google Scholar]
- 7.Kheder E.S., Hong D.S. Emerging Targeted Therapy for Tumors with NTRK Fusion Proteins. Clin. Cancer Res. 2018;24:5807–5814. doi: 10.1158/1078-0432.CCR-18-1156. [DOI] [PubMed] [Google Scholar]
- 8.Brenca M., Rossi S., Polano M., Gasparotto D., Zanatta L., Racanelli D., Valori L., Lamon S., Dei Tos A.P., Maestro R. Transcriptome sequencing identifies ETV6-NTRK3 as a gene fusion involved in GIST. J. Pathol. 2016;238:543–549. doi: 10.1002/path.4677. [DOI] [PubMed] [Google Scholar]
- 9.Shi E., Chmielecki J., Tang C.M., Wang K., Heinrich M.C., Kang G., Corless C.L., Hong D., Fero K.E., Murphy J.D., et al. FGFR1 and NTRK3 actionable alterations in “Wild-Type” gastrointestinal stromal tumors. J. Transl. Med. 2016;14:339. doi: 10.1186/s12967-016-1075-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drilon A., Laetsch T.W., Kummar S., DuBois S.G., Lassen U.N., Demetri G.D., Nathenson M., Doebele R.C., Farago A.F., Pappo A.S., et al. Efficacy of Larotrectinib in TRK Fusion-Positive Cancers in Adults and Children. N. Engl. J. Med. 2018;378:731–739. doi: 10.1056/NEJMoa1714448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doebele R.C., Drilon A., Paz-Ares L., Siena S., Shaw A.T., Farago A.F., Blakely C.M., Seto T., Cho B.C., Tosi D., et al. Entrectinib in patients with advanced or metastatic NTRK fusion-positive solid tumours: Integrated analysis of three phase 1–2 trials. Lancet Oncol. 2020;21:271–282. doi: 10.1016/S1470-2045(19)30691-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang Z., Wang L., Tang G., Medeiros L.J. Fluorescence in Situ Hybridization (FISH) for Detecting Anaplastic Lymphoma Kinase (ALK) Rearrangement in Lung Cancer: Clinically Relevant Technical Aspects. Int. J. Mol. Sci. 2019;20:3939. doi: 10.3390/ijms20163939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lange A.M., Lo H.W. Inhibiting TRK Proteins in Clinical Cancer Therapy. Cancers. 2018;10:105. doi: 10.3390/cancers10040105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pulciani S., Santos E., Lauver A.V., Long L.K., Aaronson S.A., Barbacid M. Oncogenes in solid human tumours. Nature. 1982;300:539–542. doi: 10.1038/300539a0. [DOI] [PubMed] [Google Scholar]
- 15.Vaishnavi A., Capelletti M., Le A.T., Kako S., Butaney M., Ercan D., Mahale S., Davies K.D., Aisner D.L., Pilling A.B., et al. Oncogenic and drug-sensitive NTRK1 rearrangements in lung cancer. Nat. Med. 2013;19:1469–1472. doi: 10.1038/nm.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greco A., Miranda C., Pierotti M.A. Rearrangements of NTRK1 gene in papillary thyroid carcinoma. Mol. Cell. Endocrinol. 2010;321:44–49. doi: 10.1016/j.mce.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 17.Sartore-Bianchi A., Ardini E., Bosotti R., Amatu A., Valtorta E., Somaschini A., Raddrizzani L., Palmeri L., Banfi P., Bonazzina E., et al. Sensitivity to Entrectinib Associated With a Novel LMNA-NTRK1 Gene Fusion in Metastatic Colorectal Cancer. J. Natl. Cancer Inst. 2016;108 doi: 10.1093/jnci/djv306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tognon C., Knezevich S.R., Huntsman D., Roskelley C.D., Melnyk N., Mathers J.A., Becker L., Carneiro F., MacPherson N., Horsman D., et al. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell. 2002;2:367–376. doi: 10.1016/S1535-6108(02)00180-0. [DOI] [PubMed] [Google Scholar]
- 19.Bishop J.A., Yonescu R., Batista D., Begum S., Eisele D.W., Westra W.H. Utility of mammaglobin immunohistochemistry as a proxy marker for the ETV6-NTRK3 translocation in the diagnosis of salivary mammary analogue secretory carcinoma. Hum. Pathol. 2013;44:1982–1988. doi: 10.1016/j.humpath.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cocco E., Scaltriti M., Drilon A. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat. Rev. Clin. Oncol. 2018;15:731–747. doi: 10.1038/s41571-018-0113-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaishnavi A., Le A.T., Doebele R.C. TRKing down an old oncogene in a new era of targeted therapy. Cancer Discov. 2015;5:25–34. doi: 10.1158/2159-8290.CD-14-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong D.S., DuBois S.G., Kummar S., Farago A.F., Albert C.M., Rohrberg K.S., van Tilburg C.M., Nagasubramanian R., Berlin J.D., Federman N., et al. Larotrectinib in patients with TRK fusion-positive solid tumours: A pooled analysis of three phase 1/2 clinical trials. Lancet Oncol. 2020;21:531–540. doi: 10.1016/S1470-2045(19)30856-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marchio C., Scaltriti M., Ladanyi M., Iafrate A.J., Bibeau F., Dietel M., Hechtman J.F., Troiani T., Lopez-Rios F., Douillard J.Y., et al. ESMO recommendations on the standard methods to detect NTRK fusions in daily practice and clinical research. Ann. Oncol. 2019;30:1417–1427. doi: 10.1093/annonc/mdz204. [DOI] [PubMed] [Google Scholar]
- 24.Solomon J.P., Benayed R., Hechtman J.F., Ladanyi M. Identifying patients with NTRK fusion cancer. Ann. Oncol. 2019;30((Suppl. S8)):viii16–viii22. doi: 10.1093/annonc/mdz384. [DOI] [PubMed] [Google Scholar]
- 25.Penault-Llorca F., Rudzinski E.R., Sepulveda A.R. Testing algorithm for identification of patients with TRK fusion cancer. J. Clin. Pathol. 2019;72:460–467. doi: 10.1136/jclinpath-2018-205679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiang S., Cotzia P., Hyman D.M., Drilon A., Tap W.D., Zhang L., Hechtman J.F., Frosina D., Jungbluth A.A., Murali R. NTRK fusions define a novel uterine sarcoma subtype with features of fibrosarcoma. Am. J. Surg. Pathol. 2018;42:791. doi: 10.1097/PAS.0000000000001055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lezcano C., Shoushtari A.N., Ariyan C., Hollmann T.J., Busam K.J. Primary and metastatic melanoma with NTRK-Fusions. Am. J. Surg. Pathol. 2018;42:1052. doi: 10.1097/PAS.0000000000001070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milione M., Ardini E., Christiansen J., Valtorta E., Veronese S., Bosotti R., Pellegrinelli A., Testi A., Pietrantonio F., Fucà G. Identification and characterization of a novel SCYL3-NTRK1 rearrangement in a colorectal cancer patient. Oncotarget. 2017;8:55353. doi: 10.18632/oncotarget.19512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castillon M., Kammerer-Jacquet S.F., Cariou M., Costa S., Conq G., Samaison L., Douet-Guilbert N., Marcorelles P., Doucet L., Uguen A. Fluorescent In Situ Hybridization Must be Preferred to pan-TRK Immunohistochemistry to Diagnose NTRK3-rearranged Fastrointestinal Stromal Tumors (GIST) Appl. Immunohistochem. Mol. Morphol. 2021;29:626–634. doi: 10.1097/PAI.0000000000000933. [DOI] [PubMed] [Google Scholar]
- 30.Atiq M.A., Davis J.L., Hornick J.L., Dickson B.C., Fletcher C.D.M., Fletcher J.A., Folpe A.L., Mariño-Enríquez A. Mesenchymal tumors of the gastrointestinal tract with NTRK rearrangements: A clinicopathological, immunophenotypic, and molecular study of eight cases, emphasizing their distinction from gastrointestinal stromal tumor (GIST) Mod. Pathol. 2021;34:95–103. doi: 10.1038/s41379-020-0623-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.