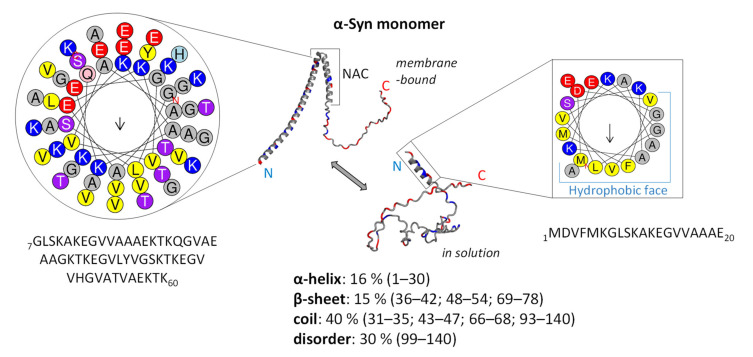
Figure 1.
Structural diversity of α-Syn. Helical conformation of α-Syn bound to a membrane and intrinsically disordered α-Syn in solution are presented in the middle. Positively charged amino acid residues are given in blue, and negative ones in red. Secondary structure propensity was obtained by predictor FELLS [63]. Helical projections of N-terminal α-helices (a total of 60 amino acid residues and the first twenty) are generated using the HeliQuest web server [64]. The arrow shows the helical hydrophobic moment. The protein structure was predicted using AlfaFold2 [65]. The 3D protein structure was visualised in PyMOL v2 (available at: https://pymol.org/2/; accessed on 16 April 2022).