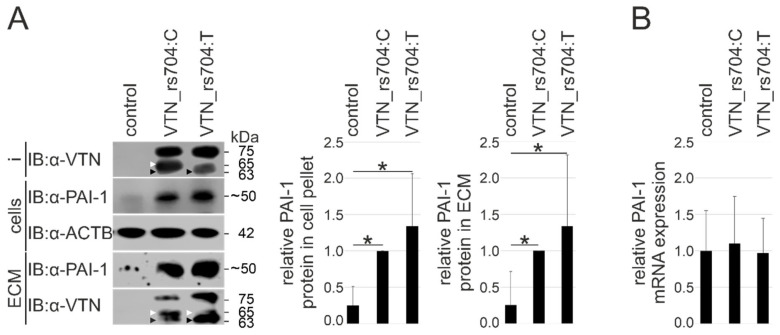
Figure 3.
Effect of vitronectin isoforms on endogenous endothelial PAI-1 protein expression and deposition. HUVECs were incubated for 24 h with input (i) including VTN_rs704: C, VTN_rs704: T, or the same volume of control eluate (control), respectively. (A) Input, cells and ECMs were subjected to immunoblot (IB) analysis with antibodies against vitronectin, PAI-1 and ACTB. Endogenous PAI-1 is recognizable as a major molecular weight species of approximately 50 kDa, consistent with previous reports of glycosylated PAI-1 as a 50–54 kDa molecular weight species [46,47]. Arrowheads indicate different fractions of recombinant vitronectin after its proteolytic cleavages (white arrows: cleavage between aa Arg398 and Ala399, black arrows: cleavage between aa Arg380 and Ser381) by endogenously expressed endoproteases. In contrast to VTN_rs704: C that stains as a mixture of a single polypeptide chain (about 75 kDa) and a clipped form (two chains of 65 and 10 kDa, held together by disulfide bonds, with the 10 kDa chain not detectable within this experimental setup), VTN_rs704: T is less susceptible to the proteolytic cleavage at Arg398-Ala399 indicated by a higher proportion of the uncleaved 75 kDa protein [8,32]. Molecular weights of approximately 63 kDa (black arrows) depict vitronectin fragments resulting from cleavage at Arg380-Ser381, as described in [50]. PAI-1 signals were densitometrically quantified and normalized against ACTB. Data represent the mean ± SD of nine biological replicates, calibrated against the PAI-1 signals obtained after VTN_rs704: C addition. Asterisks indicate statistically significant differences (* p < 0.05, Kruskal–Wallis test, followed by Dunn’s multiple comparison test with Bonferroni correction). (B) mRNA was isolated and PAI-1 expression determined by qRT-PCR. Data represent the mean ± SD of six biological replicates, calibrated against the control.