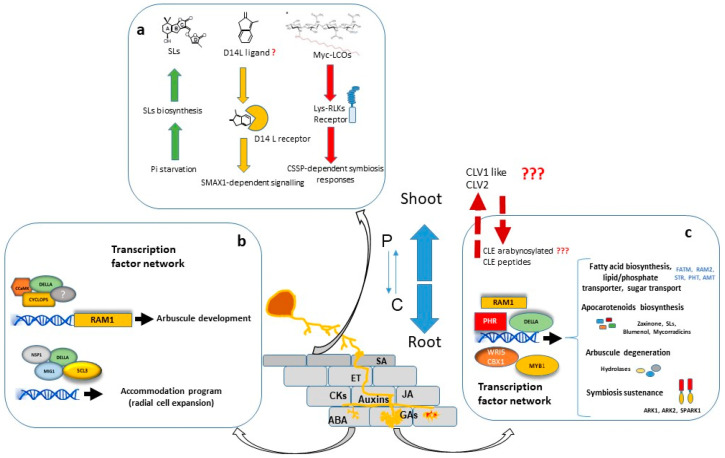
Figure 1.
Development of the arbuscular mycorrhizal (AM) symbiosis and its molecular regulation at different stages. AM symbiosis facilitates mutual nutritional benefits: the plant provides carbon (C) to AM fungi in form of carbohydrates and fatty acids, while AM fungi provide minerals, mainly inorganic phosphate (Pi) and nitrogen to the plant. The availability of the different nutrients is a key factor determining the establishment and development of the interaction. AM symbiosis alters plant hormonal homeostasis, and phytohormones play a key regulatory role in the establishment and functioning of the AM symbiosis at different levels: regulating physiology and growth, as well as defense and stress adaptation in AM plants, and participating in the formation and turnover of AM structures. At initial stages (a), Strigolactones (SLs), secreted from host roots under Pi starvation, induce fungal spore germination and hyphal branching, facilitating the contact of the fungal hypha with the root and the formation of the hyphopodium. In response to AM perception, a complex of D14L receptor with a yet unknown D14L ligand is proposed to promote the degradation of SMAX1 and the downstream activation of target genes, including symbiosis-related and SL synthesis genes necessary for the accommodation of AM fungi in host cells. AM-released chitinaceous molecules (Myc-LCOs) are proposed to be detected by specific Lys-RLKs with the formation of a receptor complex that leads to the induction of the common symbiosis signaling pathway (CSSP)-dependent symbiosis responses, including transcriptional regulation of AM symbiosis-related genes, which promotes the successful colonization of host root cells by AM fungi. A number of CSSP-activated nuclear transcription factors including CCaMK and CYCLOPS form complexes with a large number of GRAS domain-containing transcription factors, including DELLA, MIG1 and SCL3 that enable the downstream activation of symbiotic genes that facilitate the morphological accommodation of the root cell hosting the fungus, and possibility arbuscule development through the activation of RAM1 (b). Arbuscule functionality, maintenance and turnover (c) are also dependent on a number of nuclear transcription factors such as PHR, RAM1, DELLA, MYB1 and WRI5A/B that regulate transcriptional programs related to biosynthesis and the transport of nutrients, and biosynthesis of apocarotenoids compounds with regulatory roles. PHRs integrate phosphate starvation signaling, regulating mechanisms for both direct uptake of soil phosphate and AM symbiosis. DELLA and MYB1 regulate arbuscule degradation through activation of genes involved in hydrolysis processes. Receptor Kinase-2 from the subfamily of land plant-specific receptor-like kinases (rice ARK1, ARK2 and SPARK1), has been characterized as having a role in the post-arbuscule formation required for sustaining root colonization. After fungal colonization, root-derived CLE peptides (MtCLE53 and MtCLE33 in Medicago) have been proposed as activating signals that trigger a mycorrhizal autoregulation response (AOM) in shoots. The participation of CLV1-like receptors such as SUNN and the hydroxyproline O-arabinosyltransferase RDN1 required for post-translational CLE peptide modification in the AOM signaling process has been proposed.