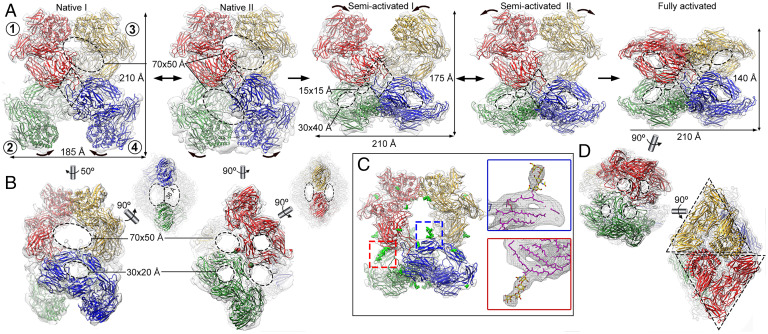
Fig. 2.
Cryo-EM structures of (hα2M)4 functional states. (A) Five functional states of (hα2M)4 from human plasma are isolated from cryo-EM analysis, termed native I (Left) and II (Center Left), semi-activated I (Center) and II (Center Right), and activated (Right). Protomer nomenclature [1 (red), 2 (green), 3 (yellow), and 4 (blue)] as described in Fig. 1. The red protomer has a disulfide-linked (green), a vicinal (yellow), and an opposite neighbor (blue). Native I and II states have protomers with expanded conformation and are in an equilibrium, at which vicinal dimers are in a distal (native I) or proximal (native II) position (curved arrows). After proteolytic activation, the native state becomes semi-activated states I and II, and one vicinal dimer is built of protomers in compact conformation. Semi-activated states I and II, which correspond to the nascent state described in the literature (46), evolve to the activated state, shown in “H-view,” in which all protomers are compact. Openings are indicated as dashed ovals. Tethering loops of opposite protomers (1, 4) are framed in a dashed rhombus. (B) Additional views of the native I (Left) and II (Right) complexes of A. The latter highlights the disulfide-linked residues between protomers 1 and 2 (red and green spheres). Dashed ellipses denote openings. (C) The semi-activated II complex highlights the N-linked glycosylation sites as green sticks (Left). Magnified view of the red and blue boxes that correspond to the same glycan bound to MG4 Asn396 in the compact protomer 4 (blue) and in the expanded protomer 1 (red). In the Insets, protein backbones are shown as magenta sticks, glycan chains as stick models with yellow carbon atoms. (D) The activated state is shown in end view (Top) and X-view (Bottom), in which the triangular prism profile for each protomer is framed. Dashed ellipses denote openings.