FIG. 3.
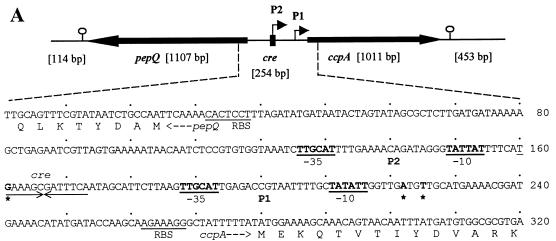
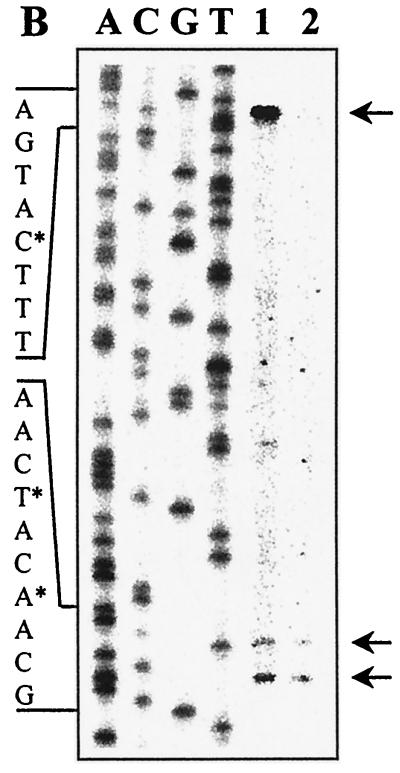
Genetic organization and transcriptional regulation of L. pentosus ccpA. (A) Genetic organization of the ccpA region. The size and orientation of the ORFs were deduced from the nucleotide sequence. The ccpA promoter region is depicted at the DNA sequence level. ccpA transcriptional start sites are in boldface and are marked by asterisks. Potential RBSs and cre motifs are underlined. Putative RNA polymerase binding sites (−10 region and −35 region) in the sequence are in boldface letters and underlined and are indicated with P1 and P2. The N-terminal protein sequences of pepQ and ccpA are shown. (B) Primer extension analysis of ccpA gene transcription of L. pentosus wild-type and ccpA mutant strains. Total RNA was prepared from cells grown on M medium supplemented with 50 mM glucose. Reverse transcription was carried out with end-labelled oligonucleotide LPE27. DNA sequencing reactions were performed with the same oligonucleotide and with pWH156 as template DNA. Primer extension products were analyzed on 6% polyacrylamide–urea gels. Lane 1, RNA (20 μg) from L. pentosus LPE4 (ccpA mutant); lane 2, RNA (20 μg) from L. pentosus MD363 (wild type). The sequence interpretations around the +1 sites (asterisks and arrows) of the two ccpA promoters are shown.