Abstract
Male sex and advanced age are associated with severe symptoms of COVID-19. Sex and age also exhibit substantial associations with genome-wide DNA methylation (DNAm) differences in humans. Using a random sample of Illumina EPIC-based genome-wide methylomes from peripheral whole blood of 1,976 parents, participating in The Norwegian Mother, Father and Child Cohort Study (MoBa), we explored whether DNAm in genes linked to SARS-CoV-2 host cell entry and to severe COVID-19 were associated with sex and age. This was carried out by testing 1,572 DNAm sites (CpGs) located near 45 genes for associations with age and sex. We found that DNAm in 281 and 231 of 1,572 CpGs were associated (pFDR<0.01) with sex and aging, respectively. CpGs linked to SARS-CoV-2 host cell entry genes were all associated with age and sex, except for the ACE2 receptor gene (located on the X-chromosome), which was only associated with sex (pFDR<0.01). Furthermore, we examined whether 1,487 autosomal CpGs associated with host-cell entry and severe COVID-19 were more or less associated with sex and age than what would be expected from the same number of randomly sampled genome-wide CpGs. We found that the CpGs associated with host-cell entry and severe COVID-19 were not more or less associated with sex (R2 = 0.77, p = 0.09) than the CpGs sampled from random genomic regions; age was actually found to be significantly less so (R2 = 0.36, p = 0.04). Hence, while we found wide-spread associations between sex and age at CpGs linked to genes implicated with SARS-CoV-2 host cell entry and severe COVID-19, the effect from the sum of these CpGs was not stronger than that from randomly sampled CpGs; for age it was significantly less so. These findings could suggest that advanced age and male sex may not be unsurmountable barriers for the SARS-CoV-2 virus to evolve increased infectiousness.
Introduction
The clinical picture of coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus, shows substantial variation; from asymptomatic to severe pneumonia-like distress requiring the aid of respirators and, in some cases, even death [1]. The SARS-CoV-1 and SARS-CoV-2 Betacoronaviruses share the same host species, i.e. horseshoe bats (Rhinolophus sp.), in addition to several genetic and phenotypic similarities [2]. In humans, both viruses bind to a receptor, coded by a gene located on the X chromosome expressing an angiotensin converting enzyme (ACE2), with a trimeric Spike protein (S) [2]. The S protein is divided into the subdomains S1, which includes the receptor binding domain (RBD), and S2 that mediates viral-host membrane fusion [2] (See S1 Fig). Current knowledge suggests that SARS-CoV-2 can enter host cells via two routes [3, 4]. One route, the early entry pathway, is through virus-host cell membrane fusion. The other described route, known as the late entry pathway, is via virus attachment to the ACE2 receptor at the cell surface and endocytosis. Both modes of entry have in common that they are currently believed to exclusively go through the ACE2 receptor [3]. For the early entry pathway, the furin site, located between the S1 and S2 subdomains of the S protein, must be cleaved before the transmembrane serine 2 protease (TMPRSS2) can cleave the S2’ site. The cleavage of the furin site takes place during virus assembly and maturation [4]. The S2’ site is located within the S2 subdomain of the S protein and becomes exposed during binding to ACE2 if the furin site is cleaved [3]. Neuropilin-1 (NRP1) has recently been found to enhance SARS-CoV-2 infectivity through the early pathway entry but is not necessary for infection [5]. Cleavage of the furin site seems to complicate the late pathway entry as the S protein is not in an optimal state for entry via endocytosis [3]. Conversely, while complicating the early pathway entry due to the required cleavage, the uncleaved furin site increases affinity for the late pathway entry through endocytosis [3]. Cleavage of the furin site leads to a less stable S protein which seems to favor early versus late pathway entry. During the endocytosis phase of the late pathway entry, cathepsin L/B (CTSL and CTSB) cleave the S’ site before the viral genome is released into the cell for subsequent viral biosynthesis [3, 4].
Host infectivity to SARS-CoV-2 depends both on viral- and host genetic factors [6]. The gene coding for the ACE2 receptor is located on the X chromosome [7]. In humans, females have two copies of the X chromosome, inherited from each parent, while males have one maternally inherited copy [8]. The second copy of the X chromosome in females is replaced by a paternally inherited Y chromosome in males [9]. One of the two X chromosomes located in female cells is inactivated (XCI) which is marked by ubiquitous DNA methylation (DNAm) [8]. DNAm is typically taken to be the addition of a methyl group to Cytosine in Cytosine-Guanine (CpG) di-nucleotide pairs [10]. Which of the two X-chromosomes in females become inactivated varies randomly from cell to cell [8]. Hence, female cells express one of two ACE2 receptor variants inherited from each parents while males express only a maternally inherited variant [11, 12]. Due to the strong association with cell type differentiation [13], DNAm is the most studied epigenetic mark [10]. It has also been found that genome-wide DNAm varies throughout the life-course and as a consequence of genetics and environmental exposures [14]. In particular, a substantial fraction of genome-wide DNAm differences have both been attributed to age and sex differences in humans [15, 16]. Indeed, numerous DNAm-based epigenetic clocks have been created that can predict chronological age from different cell types [17]. Epigenetic clocks have also been discovered from certain cell types that can predict age with a high accuracy [18]. There are also epigenetic clocks that can measure biological age linked to all-cause mortality [19]. DNAm-based epigenetic clocks have recently been linked to T and NK cell activation suggesting that epigenetic clocks are in general reflective of host immune system status [20].
Current risk factors for severe COVID-19 symptoms are advanced age [21, 22] and being male [11]. COVID-19 outcomes may also be influenced by DNAm in multiple tissues and organs owing to the systemic nature of the disease [23]. Two recent genome-wide association studies (GWASs) of severe COVID-19 [1, 24] reported single nucleotide polymorphisms (SNPs) significantly linked to 16 genes in total. The strong association of severe COVID-19 with sex [11] and age [25] motivated us to examine whether DNAm differences associated with sex and age could be linked to genes implicated with severe COVID-19, defined as critical respiratory failure [1, 24]. We additionally scrutinized age and sex effects of CpGs related to genes involved in SARS-CoV-2 host cell entry. In particular, we examined both ACE2 and TMPRSS2 genes and their associated STRING network clusters [26] (11 + 11 = 22 genes in total) as well as 7 genes [3, 5, 27, 28] implicated with both early and late pathway entry.
In total, we looked for sex and age related DNAm differences in 1,572 CpG loci (1,487 autosomal CpGs and 85 X chromome CpGs), linked to 45 genes (41 autosomal genes and 4 X chromosome genes) associated with severe COVID-19 and SARS-CoV-2 host cell entry, in 1,976 parents participating in the Norwegian Mother, Father and Child Cohort Study (MoBa).
Materials and methods
The study population was taken from The Norwegian Mother, Father and Child Study cohort (MoBa) [29]. Information regarding the study population can be found in Table 1.
Table 1. Study population characteristics.
| Males | Females | |
|---|---|---|
| Number | 991(50.2%) | 985(49.8%) |
| Median age in years (mean) | 32.3(32.8)(18.3, 58.6) | 30.1(30.1)(18.3, 45.5) |
The table shows the study population’s sex (number (percent)) and age (median (mean)(min,max)) characteristics.
The establishment of MoBa and initial data collection was based on a license from the Norwegian Data Protection Agency and approval from The Regional Committees for Medical and Health Research Ethics (REK). The MoBa cohort is currently regulated by the Norwegian Health Registry Act. The current study was approved by REK South-East (2017/1362) in Norway 26.09.2017. Data collection by MoBa was carried out during 2001–2009 in accordance with the Norwegian Data Protection Agency after securing approval from REK. All participants provided written informed consent.
We used 1,976 randomly sampled adults (see Table 1) from MoBa [29]. The DNA methylation of these subjects were measured from peripheral blood using Illumina’s Infinium MethylationEPIC array. Quality control and preprocessing was carried out using the RnBeads package [30]. During the quality control procedure, probes with cross-hybridization, high detection P-value (>0.01) and those near single-nucleotide polymorphisms (SNP) (three last bases of the probe sequence overlaps with a SNP) were excluded resulting in a total of 770,586 out of 846,232 autosomal probes (total probes on the Illumina EPIC array is 865,859 probes, 19,090 probes are located on the X chromosome and 537 on the Y chromosome). Signal intensities were background-corrected using enmix.oob [31] before probe Type I and Type II calibrations were carried out using the beta-mixture quantile normalization (BMIQ) procedure [32]. The X chromosomes were preprocessed independently of the autosomes to prevent bias due to X chromosome inactivation. The same guidelines were used for the X chromosomes as for the autosomes. Using the same exclusion principles as for autosomes we ended up with 16,841 probes on the X chromosome on which BMIQ was performed. Further details regarding quality control of these samples can be found in Lee et al. [33]. Estimates of the different cell types were carried out using the reference set described by Salas et al. [34] using the Houseman procedure [35]. All cell types, i.e. CD4+ T and CD8+ T cells, NK cells, B cells, monocytes and neutrophils, were adjusted for in the regression analyses.
Statistical modeling was carried out using mixed-effects regression with the R package ‘nlme’ [36, 37]. DNAm at each CpG was treated as the outcome (β values), with sex and age as explanatory variables. EPIC array plates were included as random intercept effects. Regression models used to assess the DNAm effects of sex and age were reciprocally adjusted for age and sex together with cell type proportion estimates.
Adjustment for multiple testing was performed using false discovery rate (FDR) and Bonferroni on 770,586 autosomal + 16,841 X chromosomal CpG sites = 787,427 tests in total. We considered only genome-wide pFDR<0.01 as statistically significant. Analyses of CpGs located on the X-chromosome (particularly CpGs linked to the ACE2 receptor gene) were stratified for males and females due to XCI.
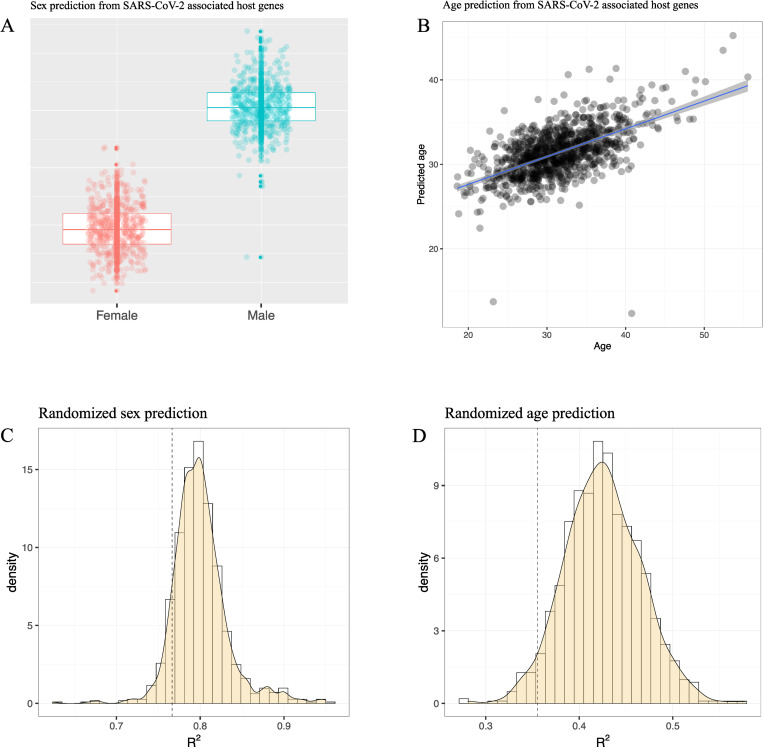
Prediction models were performed using Lasso penalized regression from the ‘glmnet’ package [38] with the X matrix consisting of the 1,487 CpGs linked to 41 genes associated with severe COVID-19 and SARS-CoV-2 host cell entry located on the autosomes. The outcome of the Lasso regression was binomial for sex (male/female) and continuous for age (days, subsequently converted to years). Half the individuals (n = 988) were selected randomly with the corresponding outcome (sex/age) for training while prediction was carried out on the other half (n = 988 individuals). Subsequent linear regressions were performed on predicted versus given age in days and sex respectively, from which the coefficient of variation R2 was recorded. For randomization, 1,487 CpGs were selected randomly from the autosomal CpGs with training and prediction performed on the same halves of the samples as above. The randomization procedure was repeated 1,000 times for both sex and age outcomes, respectively, with subsequent linear regressions on given and predicted age/sex. The variance explained parameter (R2) was recorded for the regression models and corresponding distributions were made as can be seen in Fig 2C and 2D for sex and age. The p values for R2 were computed using the empirical distributions shown in Fig 2 panels C and D. All estimations and regression results can be found in S1 Appendix.
Fig 2. Predicting sex and age in SARS-CoV-2 associated host genes.
Panel A shows the association between autosomal CpGs, linked to SARS-CoV-2 host genes/severe COVID-19, and predicted sex (vertical axis). Panel B shows the association between CpGs predicting age (vertical axis) and given age (horizontal axis). The distributions of the variance explained parameter R2, stemming from 1,000 randomizations of genome-wide randomly selected CpGs against sex and age, are shown in Panels C and D, respectively. The vertical dashed line designates the corresponding R2 for the sex (R2 = 0.77, p = 0.09) and age (R2 = 0.36, p = 0.04) regression models depicted in panels A and B.
Results and discussion
In total, we examined 1,572 CpGs (including 85 CpGs on the X chromosome) related to 45 host genes (4 genes were located on the X chromosome). Because of X chromosome inactivation (XCI) [8], we analyzed genes located on autosomes separately from genes on the X chromosome. Results from the regression analyses can be found in Table 2 as well as in S1 Appendix, which also includes regression estimates and information regarding the genes included in the study.
Table 2. SARS-CoV-2 genes associated with host cell entry and severe COVID-19.
The table shows information regarding genes associated with SARS-CoV-2 host cell entry as well as severe COVID-19. Source describes the source justifying the inclusion of the gene, Gene identifier refers to the gene symbol, Sex associated shows number of DMPs significantly associated with sex (pFDR<0.01), Age associated shows the same for age, CpGs linked to gene indicates the number of CpGs on the EPIC array linked to that particular gene and, finally, the last column Chromosome displays the chromosome number on which the gene is located.
| Source | Gene | Sex associated | Age associated | CpGs linked to gene | Chromosome |
|---|---|---|---|---|---|
| ACE2 Cluster [12] | ACE2* | 12 | 0 | 15 | X |
| AGT | 0 | 1 | 13 | 1 | |
| AGTR1 | 6 | 12 | 29 | 3 | |
| AGTR2* | 5 | 0 | 5 | X | |
| DPP4 | 6 | 7 | 36 | 2 | |
| MEP1A | 5 | 1 | 12 | 6 | |
| MEP1B | 2 | 1 | 3 | 18 | |
| MME | 2 | 16 | 35 | 3 | |
| PRCP | 9 | 1 | 38 | 11 | |
| REN | 2 | 1 | 13 | 1 | |
| XPNPEP2* | 17 | 0 | 18 | X | |
| TMPRSS2 Cluster [12] | TMPRSS2 | 1 | 7 | 32 | 21 |
| AR* | 47 | 0 | 47 | X | |
| ERG | 16 | 15 | 78 | 21 | |
| ETV1 | 10 | 6 | 45 | 7 | |
| ETV4 | 0 | 5 | 32 | 17 | |
| FAM3B | 1 | 7 | 18 | 21 | |
| FKBP5 | 9 | 5 | 49 | 6 | |
| NKX3-1 | 2 | 4 | 15 | 8 | |
| PTEN | 14 | 2 | 76 | 10 | |
| SLC45A3 | 6 | 4 | 34 | 1 | |
| TMPRSS4 | 3 | 10 | 44 | 11 | |
| GWAS [1] | ABO | 0 | 2 | 18 | 9 |
| Entry [14] | ADAM17 | 8 | 0 | 33 | 2 |
| GWAS [11] | CCHCR1 | 5 | 2 | 59 | 6 |
| GWAS [1] | CCR9 | 1 | 4 | 15 | 3 |
| Entry (late pathway) [3] | CTSB | 6 | 4 | 38 | 8 |
| Entry (late pathway) [3] | CTSL | 0 | 3 | 5 | 9 |
| GWAS [1] | CXCR6 | 2 | 1 | 14 | 3 |
| GWAS [11] | DPP9 | 5 | 1 | 52 | 19 |
| Entry (early pathway) [3] | FURIN | 7 | 5 | 47 | 15 |
| GWAS [1] | FYCO1 | 5 | 4 | 38 | 3 |
| GWAS [11] | HLA-G | 6 | 8 | 18 | 6 |
| Entry [13] | HMGB1 | 5 | 2 | 22 | 13 |
| GWAS [1] | IFNAR2 | 2 | 1 | 29 | 21 |
| GWAS [1,11] | LZTFL1 | 6 | 7 | 40 | 3 |
| GWAS [11] | NOTCH4 | 18 | 38 | 139 | 6 |
| Entry [4] | NRP1 | 13 | 14 | 79 | 10 |
| Entry [4] | NRP2 | 4 | 19 | 85 | 2 |
| GWAS [11] | OAS1 | 5 | 2 | 15 | 12 |
| GWAS [11] | OAS2 | 1 | 1 | 23 | 12 |
| GWAS [11] | OAS3 | 3 | 0 | 21 | 12 |
| GWAS [1] | SLC6A20 | 1 | 7 | 39 | 3 |
| GWAS [11] | TYK2 | 2 | 1 | 46 | 19 |
| GWAS [1] | XCR1 | 1 | 0 | 10 | 3 |
| Total | 45 | 281 (200 without X chromosome genes) | 231 | 1572 (1487 on autosomes + 85 on X chromosome) | |
‘*’ Gene located on X chromosome.
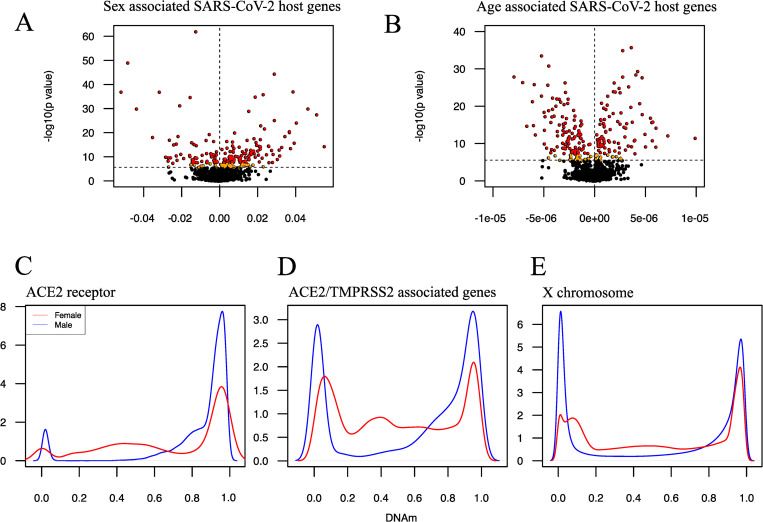
We found that 200 and 231 differentially methylated probes (DMPs), located near genes related to both SARS-CoV-2 host cell entry and severe COVID-19, were significantly associated (pFDR<0.01) with sex and age respectively on autosomes (See Fig 1, panels A and B). While sex-associated CpGs (Fig 1A) were more hyper-methylated (800 vs 687 CpGs, 133 vs 69 DMPs pFDR<0.01) age-associated CpGs (Fig 1B) were predominantly hypo-methylated (839 vs 648 CpGs, 130 vs 101 DMPs pFDR<0.01). Linear regressions of DNAm of CpGs on the X-chromosome, assessing whether sex differences could be found at any of the sites connected to the genes linked with SARS-CoV-2 host cell entry (no genes currently identified with severe COVID-19 were located on the X chromosome), resulted in 81 significant DMPs (pFDR<0.01) out of 85. In general, there are profound DNAm differences between males and females on the X chromosome (Fig 1 panels C, D and E). Whether these are true differences or artifacts on the EPIC array resulting from XCI is unclear. We therefore performed stratified regression analysis for males and females with age as the explanatory variable and DNAm from each CpG as outcome (β values). Age was not significantly associated with any CpGs related to the ACE2 gene or any other gene linked to SARS-CoV-2 host cell entry on the X chromosome. This may suggest that the SARS-CoV-2 virus may bind with equal affinity to the ACE2 host receptor in all age groups in peripheral blood. The CpGs on the EPIC array located near the ACE2 gene are mainly found in transcription start sites (TSSs) (5 CpGs), untranslated regions (UTR’s)(2 CpGs), gene bodies (6 CpGs) and exons (2 CpGs) according to UCSC annotation (https://genome.ucsc.edu/). The five CpGs associated with the ACE2 gene annotated to TSS were hyper-methylated, with some slight hemi-methylation in females that could be an artifact of XCI (see S2 Fig). This could indicate that the ACE2 gene is not expressed in whole blood, which has been suggested elsewhere [39]. If, on the other hand, the ACE2 gene is expressed, our findings may point to that the ACE2 gene is differently methylated between males and females supporting the previously reported sex differences with regards to COVID-19 infection [11]. In the ACE2 gene STRING network cluster we also find the DPP4 receptor gene for the more deadly but far less infectious Middle Eastern Respiratory Syndrome Coronavirus (MERS-CoV) [2]. For the DPP4 gene, 6 CpGs were significantly associated with sex and 7 with age. In contrast to the ACE2 receptor, the TMPRSS2 protease, linked to early pathway entry, is mostly associated with age (6 DMPs versus 1 DMP for sex). The furin protease had 7 DMPs associated with sex and 5 with age. For the genes coding for the proteases facilitating late entry pathway (CTSL and CTSB) we found that no CpGs were significantly associated with sex for CTSL, while 3 (of 5) CpGs were associated with age. For CTSB, 6 CpGs were significantly associated with sex and 4 with age. Other genes associated with SARS-CoV-2 host cell entry included metallopeptidase domain 17 (ADAM17) [28] (8 and 0 CpGs significantly associated with sex and age, respectively), high mobility group box 1 gene (HMGB1) [27] (5 and 2 DMPs), NRP1 (13 and 14 DMPs) as well as NRP2 genes [5] (4 and 19 DMPs). These findings may suggest that age could potentially influence the mode of entry for SARS-CoV-2 [40]. In particular, as the CpGs linked to the TMPRSS2 gene were strongly associated with age our findings may suggest that enhanced late pathway entry via endocytosis could increase COVID-19 infectivity by allowing additional age groups to be more susceptible to infection [41].
Fig 1. DNA methylation in genes associated with COVID-19.
Panel A) shows a volcano plot of the–log10 transformed p values (vertical axis) of the coefficients resulting from regressing DNAm in autosomal CpGs linked to COVID-19 genes on sex (horizontal axis). Black colored dots signify non-significantly associated CpGs, orange dots pFDR<0.01 significantly associated CpGs and red dots Bonferroni pB<0.05 significantly associated CpGs. Panel B is similar to Panel A but the coefficients now reflect age instead of sex. The DNAm density of the CpGs linked to the ACE2 receptor gene on the X chromosome can be seen in Panel C. Red colored lines designate females while blue lines males. Panel D shows the DNAm density for all CpGs related to the four ACE2/TMPRSS2 STRING network clustered genes (ACE2, AGTR2, AR, XPNPEP2) located on the X chromosome, while the total DNAm density differences between males and females stemming from the 16,841 X-linked CpGs is depicted in Panel E.
Only one of 16 genes were shared between the two previously mentioned GWASs: Leucine zipper transcription factor-like proten 1 (LZTFL1). Of the DMPs linked to the LZTFL1 gene, there were 6 CpGs significantly associated with sex and 7 with age. In total, we found that 63 CpGs were significantly associated with sex while 79 were associated with age of the 576 CpGs linked to the 16 genes associated with severe COVID-19.
Since it has been found that both sex and age DMPs are wide-spread in the human genome [15], we made sex- and age predictors based only on the 1,487 CpGs linked to the autosomal genes associated with SARS-CoV-2 host cell entry and severe COVID-19. We randomly selected half the samples for training of the Lasso classifier (n = 988 samples) and the other half (again n = 988 samples) for prediction. Fig 2, panels A and B, show the results from these prediction models for sex and age. The variance explained (R2) was 77% for the sex-based model and 36% for age. We also compared our Lasso models with corresponding sex- and age based models trained on 1,487 randomly selected genome-wide CpGs. These predictions were performed 1,000 times for both sex and age models and R2 distributions from the subsequent regression models can be seen in Fig 2C and 2D, respectively. Fig 2C indicates that the prediction model for sex did not perform significantly different, with respect to R2, from genome-wide random draws of CpGs (p = 0.09). For age (Fig 2D), we found that R2 was actually slightly lower than expected compared to R2 from randomly drawn CpGs (p = 0.04). Hence, while we found several sex and age related DMPs linked to genes associated with severe COVID-19 and SARS-CoV-2 host cell entry the proportion did not exceed what would be expected from random genome-wide sampling of CpGs. While we found no differences for sex related DMPs linked to severe COVID-19 and host cell entry connected genes, for age related DMPs the effect was found to be lower than expected. Our findings therefore may suggest that age and sex may not be impossible barriers for the SARS-CoV-2 virus to increase infectiousness [42].
Strengths of our study include the use of a large randomly selected and homogeneous population as well as the use of the Illumina EPIC array with 850K probes. A weakness of this study is the restriction to peripheral blood. A recent study did however find strong associations between DNAm and COVID-19 infection in peripheral blood [43]. Nevertheless, scrutiny of CpGs in cells from the upper and lower respiratory tract is likely to be more revealing in terms of SARS-CoV-2 host cell entry. Another more general weakness is that the effects reported here are likely underestimations as the EPIC array only covers approximately 3% of methylation sites in the human genome [44].
Conclusions
In summary, we found no association between age and DMPs related to the ACE2 receptor gene but several DMPs associated with sex. Whether these differences are due to X inactivation bias is not known. Furthermore, we found no hypo-methylated DMPs linked to the ACE2 TSS, raising the question whether this receptor is at all expressed in peripheral blood. In general, we detected widespread associations between sex and age in CpGs linked to genes associated with host cell entry as well as severe COVID-19. However, these DMPs did not appear to be more strongly associated with sex and age than CpGs drawn genome-wide at random suggesting that age and sex may not be impossible obstacles for the SARS-CoV-2 virus to surmount to increase infectivity.
Supporting information
(DOCX)
(PDF)
(XLSX)
Acknowledgments
MoBa is supported by the Norwegian Ministry of Health and Care Services and the Ministry of Education and Research. We are deeply grateful to all the families in Norway who participate in this ongoing cohort study. The Illumina EPIC arrays were processed at Life and Brain GmbH in Bonn, Germany (www.lifeandbrain.com). All analyses were performed on the HUNT Cloud service at the Norwegian University of Science and Technology (NTNU) for sensitive data.
Data Availability
The MoBa data can be accessed by applying directly to the Norwegian Institute of Public Health, https://www.fhi.no/en/studies/moba/.
Funding Statement
The START project was funded by the Research Council of Norway through its Centres of Excellence funding scheme, project number 262700, and the NIPH. JHOP is funded by the Swedish research council FORMAS (grant no: 2015-710) and VR (grant no: 2020-02593). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Severe Covid GG, Ellinghaus D, Degenhardt F, Bujanda L, Buti M, Albillos A, et al. Genomewide Association Study of Severe Covid-19 with Respiratory Failure. N Engl J Med. 2020;383(16):1522–34. Epub 2020/06/20. doi: 10.1056/NEJMoa2020283 ; PubMed Central PMCID: PMC7315890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berrio A, Gartner V, Wray GA. Positive selection within the genomes of SARS-CoV-2 and other Coronaviruses independent of impact on protein function. PeerJ. 2020;8:e10234. Epub 2020/10/23. doi: 10.7717/peerj.10234 ; PubMed Central PMCID: PMC7571416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang T, Jaimes JA, Bidon MK, Straus MR, Daniel S, Whittaker GR. Proteolytic Activation of SARS-CoV-2 Spike at the S1/S2 Boundary: Potential Role of Proteases beyond Furin. ACS Infect Dis. 2021;7(2):264–72. Epub 2021/01/13. doi: 10.1021/acsinfecdis.0c00701 ; PubMed Central PMCID: PMC7839419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jackson CB, Farzan M, Chen B, Choe H. Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol. 2022;23(1):3–20. Epub 2021/10/07. doi: 10.1038/s41580-021-00418-x ; PubMed Central PMCID: PMC8491763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cantuti-Castelvetri L, Ojha R, Pedro LD, Djannatian M, Franz J, Kuivanen S, et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370(6518):856–60. Epub 2020/10/22. doi: 10.1126/science.abd2985 ; PubMed Central PMCID: PMC7857391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19(7):409–24. Epub 2021/06/03. doi: 10.1038/s41579-021-00573-0 ; PubMed Central PMCID: PMC8167834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou H, Chen X, Hu T, Li J, Song H, Liu Y, et al. A Novel Bat Coronavirus Closely Related to SARS-CoV-2 Contains Natural Insertions at the S1/S2 Cleavage Site of the Spike Protein. Curr Biol. 2020;30(11):2196–203 e3. Epub 2020/05/18. doi: 10.1016/j.cub.2020.05.023 ; PubMed Central PMCID: PMC7211627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li S, Lund JB, Christensen K, Baumbach J, Mengel-From J, Kruse T, et al. Exploratory analysis of age and sex dependent DNA methylation patterns on the X-chromosome in whole blood samples. Genome Med. 2020;12(1):39. Epub 2020/04/30. doi: 10.1186/s13073-020-00736-3 ; PubMed Central PMCID: PMC7189689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lund JB, Li S, Christensen K, Mengel-From J, Soerensen M, Marioni RE, et al. Age-dependent DNA methylation patterns on the Y chromosome in elderly males. Aging Cell. 2020;19(2):e12907. Epub 2019/02/23. doi: 10.1111/acel.12907 ; PubMed Central PMCID: PMC6996942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carmona JJ, Accomando WP Jr., Binder AM, Hutchinson JN, Pantano L, Izzi B, et al. Empirical comparison of reduced representation bisulfite sequencing and Infinium BeadChip reproducibility and coverage of DNA methylation in humans. NPJ Genom Med. 2017;2:13. Epub 2017/12/22. doi: 10.1038/s41525-017-0012-9 ; PubMed Central PMCID: PMC5642382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peckham H, de Gruijter NM, Raine C, Radziszewska A, Ciurtin C, Wedderburn LR, et al. Male sex identified by global COVID-19 meta-analysis as a risk factor for death and ITU admission. Nat Commun. 2020;11(1):6317. Epub 2020/12/11. doi: 10.1038/s41467-020-19741-6 ; PubMed Central PMCID: PMC7726563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beacon TH, Delcuve GP, Davie JR. Epigenetic regulation of ACE2, the receptor of the SARS-CoV-2 virus(1). Genome. 2021;64(4):386–99. Epub 2020/10/22. doi: 10.1139/gen-2020-0124 . [DOI] [PubMed] [Google Scholar]
- 13.Scott CA, Duryea JD, MacKay H, Baker MS, Laritsky E, Gunasekara CJ, et al. Identification of cell type-specific methylation signals in bulk whole genome bisulfite sequencing data. Genome Biol. 2020;21(1):156. Epub 2020/07/02. doi: 10.1186/s13059-020-02065-5 ; PubMed Central PMCID: PMC7329512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Dongen J, Nivard MG, Willemsen G, Hottenga JJ, Helmer Q, Dolan CV, et al. Genetic and environmental influences interact with age and sex in shaping the human methylome. Nat Commun. 2016;7:11115. Epub 2016/04/08. doi: 10.1038/ncomms11115 ; PubMed Central PMCID: PMC4820961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yusipov I, Bacalini MG, Kalyakulina A, Krivonosov M, Pirazzini C, Gensous N, et al. Age-related DNA methylation changes are sex-specific: a comprehensive assessment. Aging (Albany NY). 2020;12(23):24057–80. Epub 2020/12/05. doi: 10.18632/aging.202251 ; PubMed Central PMCID: PMC7762479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singmann P, Shem-Tov D, Wahl S, Grallert H, Fiorito G, Shin SY, et al. Characterization of whole-genome autosomal differences of DNA methylation between men and women. Epigenetics Chromatin. 2015;8:43. Epub 2015/10/27. doi: 10.1186/s13072-015-0035-3 ; PubMed Central PMCID: PMC4615866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115. Epub 2013/10/22. doi: 10.1186/gb-2013-14-10-r115 ; PubMed Central PMCID: PMC4015143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horvath S, Oshima J, Martin GM, Lu AT, Quach A, Cohen H, et al. Epigenetic clock for skin and blood cells applied to Hutchinson Gilford Progeria Syndrome and ex vivo studies. Aging (Albany NY). 2018;10(7):1758–75. Epub 2018/07/27. doi: 10.18632/aging.101508 ; PubMed Central PMCID: PMC6075434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marioni RE, Shah S, McRae AF, Chen BH, Colicino E, Harris SE, et al. DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol. 2015;16:25. Epub 2015/01/31. doi: 10.1186/s13059-015-0584-6 ; PubMed Central PMCID: PMC4350614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jonkman TH, Dekkers KF, Slieker RC, Grant CD, Ikram MA, van Greevenbroek MMJ, et al. Functional genomics analysis identifies T and NK cell activation as a driver of epigenetic clock progression. Genome Biol. 2022;23(1):24. Epub 2022/01/16. doi: 10.1186/s13059-021-02585-8 ; PubMed Central PMCID: PMC8759260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Atlante S, Mongelli A, Barbi V, Martelli F, Farsetti A, Gaetano C. The epigenetic implication in coronavirus infection and therapy. Clin Epigenetics. 2020;12(1):156. Epub 2020/10/23. doi: 10.1186/s13148-020-00946-x ; PubMed Central PMCID: PMC7576975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El Baba R, Herbein G. Management of epigenomic networks entailed in coronavirus infections and COVID-19. Clin Epigenetics. 2020;12(1):118. Epub 2020/08/08. doi: 10.1186/s13148-020-00912-7 ; PubMed Central PMCID: PMC7404079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corley MJ, Pang APS, Dody K, Mudd PA, Patterson BK, Seethamraju H, et al. Genome-wide DNA methylation profiling of peripheral blood reveals an epigenetic signature associated with severe COVID-19. J Leukoc Biol. 2021. Epub 2021/01/20. doi: 10.1002/JLB.5HI0720-466R ; PubMed Central PMCID: PMC8013321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pairo-Castineira E, Clohisey S, Klaric L, Bretherick AD, Rawlik K, Pasko D, et al. Genetic mechanisms of critical illness in COVID-19. Nature. 2021;591(7848):92–8. Epub 2020/12/12. doi: 10.1038/s41586-020-03065-y . [DOI] [PubMed] [Google Scholar]
- 25.Rydyznski Moderbacher C, Ramirez SI, Dan JM, Grifoni A, Hastie KM, Weiskopf D, et al. Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity. Cell. 2020;183(4):996–1012 e19. Epub 2020/10/05. doi: 10.1016/j.cell.2020.09.038 ; PubMed Central PMCID: PMC7494270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, et al. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37(Database issue):D412–6. Epub 2008/10/23. doi: 10.1093/nar/gkn760 ; PubMed Central PMCID: PMC2686466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei J, Alfajaro MM, DeWeirdt PC, Hanna RE, Lu-Culligan WJ, Cai WL, et al. Genome-wide CRISPR Screens Reveal Host Factors Critical for SARS-CoV-2 Infection. Cell. 2021;184(1):76–91 e13. Epub 2020/11/05. doi: 10.1016/j.cell.2020.10.028 ; PubMed Central PMCID: PMC7574718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coate KC, Cha J, Shrestha S, Wang W, Goncalves LM, Almaca J, et al. SARS-CoV-2 Cell Entry Factors ACE2 and TMPRSS2 Are Expressed in the Microvasculature and Ducts of Human Pancreas but Are Not Enriched in beta Cells. Cell Metab. 2020;32(6):1028–40 e4. Epub 2020/11/19. doi: 10.1016/j.cmet.2020.11.006 ; PubMed Central PMCID: PMC7664344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Magnus P, Birke C, Vejrup K, Haugan A, Alsaker E, Daltveit AK, et al. Cohort Profile Update: The Norwegian Mother and Child Cohort Study (MoBa). Int J Epidemiol. 2016;45(2):382–8. Epub 2016/04/12. doi: 10.1093/ije/dyw029 . [DOI] [PubMed] [Google Scholar]
- 30.Muller F, Scherer M, Assenov Y, Lutsik P, Walter J, Lengauer T, et al. RnBeads 2.0: comprehensive analysis of DNA methylation data. Genome Biol. 2019;20(1):55. Epub 2019/03/16. doi: 10.1186/s13059-019-1664-9 ; PubMed Central PMCID: PMC6419383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Z, Niu L, Li L, Taylor JA. ENmix: a novel background correction method for Illumina HumanMethylation450 BeadChip. Nucleic Acids Res. 2016;44(3):e20. Epub 2015/09/19. doi: 10.1093/nar/gkv907 ; PubMed Central PMCID: PMC4756845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D, et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29(2):189–96. Epub 2012/11/24. doi: 10.1093/bioinformatics/bts680 ; PubMed Central PMCID: PMC3546795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee Y, Haftorn KL, Denault WRP, Nustad HE, Page CM, Lyle R, et al. Blood-based epigenetic estimators of chronological age in human adults using DNA methylation data from the Illumina MethylationEPIC array. BMC Genomics. 2020;21(1):747. Epub 2020/10/29. doi: 10.1186/s12864-020-07168-8 ; PubMed Central PMCID: PMC7590728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salas LA, Koestler DC, Butler RA, Hansen HM, Wiencke JK, Kelsey KT, et al. An optimized library for reference-based deconvolution of whole-blood biospecimens assayed using the Illumina HumanMethylationEPIC BeadArray. Genome Biol. 2018;19(1):64. Epub 2018/05/31. doi: 10.1186/s13059-018-1448-7 ; PubMed Central PMCID: PMC5975716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. Epub 2012/05/10. doi: 10.1186/1471-2105-13-86 ; PubMed Central PMCID: PMC3532182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Team RC. R: A language and environment for statistical computing. 2013. [Google Scholar]
- 37.Pinheiro J, Bates D, DebRoy S, Sarkar D, Team RC. nlme: Linear and nonlinear mixed effects models. R package version. 2006;3(4):109. [Google Scholar]
- 38.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. Journal of statistical software. 2010;33(1):1. [PMC free article] [PubMed] [Google Scholar]
- 39.Song X, Hu W, Yu H, Zhao L, Zhao Y, Zhao X, et al. Little to no expression of angiotensin-converting enzyme-2 on most human peripheral blood immune cells but highly expressed on tissue macrophages. Cytometry A. 2020. Epub 2020/12/07. doi: 10.1002/cyto.a.24285 . [DOI] [PubMed] [Google Scholar]
- 40.Zhao H, Lu L, Peng Z, Chen LL, Meng X, Zhang C, et al. SARS-CoV-2 Omicron variant shows less efficient replication and fusion activity when compared with Delta variant in TMPRSS2-expressed cells. Emerg Microbes Infect. 2022;11(1):277–83. Epub 2021/12/25. doi: 10.1080/22221751.2021.2023329 ; PubMed Central PMCID: PMC8774049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Belay ED, Godfred-Cato S. SARS-CoV-2 spread and hospitalisations in paediatric patients during the omicron surge. Lancet Child Adolesc Health. 2022. Epub 2022/02/22. doi: 10.1016/S2352-4642(22)00060-8 ; PubMed Central PMCID: PMC8856665 are those of the authors and do not necessarily represent the official position of the US Centres for Disease Control and Prevention. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar A, Parashar R, Kumar S, Faiq MA, Kumari C, Kulandhasamy M, et al. Emerging SARS-CoV-2 variants can potentially break set epidemiological barriers in COVID-19. J Med Virol. 2022;94(4):1300–14. Epub 2021/11/24. doi: 10.1002/jmv.27467 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Balnis J, Madrid A, Hogan KJ, Drake LA, Chieng HC, Tiwari A, et al. Blood DNA methylation and COVID-19 outcomes. Clin Epigenetics. 2021;13(1):118. Epub 2021/05/27. doi: 10.1186/s13148-021-01102-9 ; PubMed Central PMCID: PMC8148415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heiss JA, Brennan KJ, Baccarelli AA, Tellez-Rojo MM, Estrada-Gutierrez G, Wright RO, et al. Battle of epigenetic proportions: comparing Illumina’s EPIC methylation microarrays and TruSeq targeted bisulfite sequencing. Epigenetics. 2020;15(1–2):174–82. Epub 2019/09/21. doi: 10.1080/15592294.2019.1656159 ; PubMed Central PMCID: PMC6961683. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(PDF)
(XLSX)
Data Availability Statement
The MoBa data can be accessed by applying directly to the Norwegian Institute of Public Health, https://www.fhi.no/en/studies/moba/.