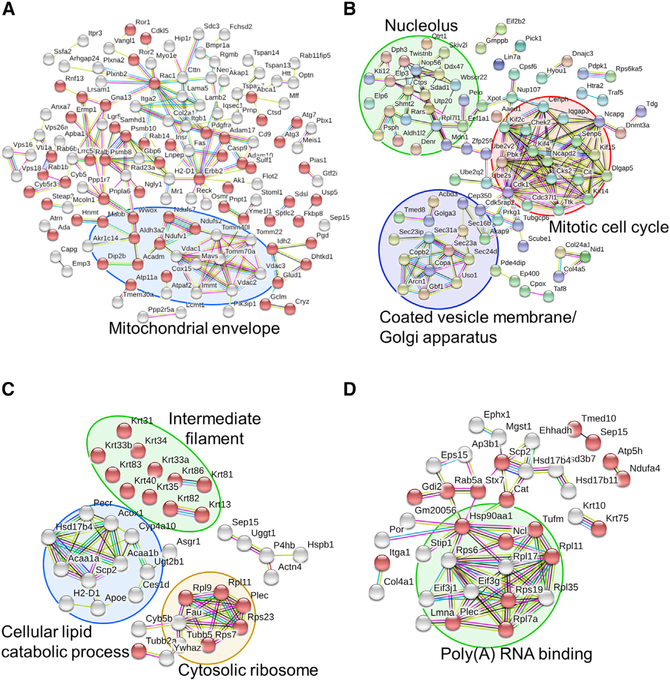
Figure 6. Network Analysis of Enriched Pathways and Interactions.
(A and B) WT and Selenof KO MEFs were cultured in serum-free media and subjected to quantitative proteomics analysis. Enriched proteins (p < 0.05, 138 up- and 198 down-regulated in Selenof KO) were further analyzed and visualized with STRING. Proteins with catalytic activity are marked with red bubbles. The functions “membrane part” and “regulation of cellular localization” were the most decreased in Selenof KO cells. Mitochondrial envelope proteins were decreased in Selenof KO (A). Proteins with increased levels in Selenof KO MEFs belonged to the clusters “Coated vesicle membrane/Golgi apparatus” (circled in purple), “Nucleolus” (circled in green), and “Mitotic cell cycle” (circled in red) (B).
(C and D) Selenof and UGGT1 interacting proteins and the associated enriched pathways. Proteins from WT and Selenof KO livers were immunoprecipitated with antibodies against Selenof (C) or UGGT1 (D), followed by MS/MS analysis. Proteins identified (p < 0.05) were furtheranalyzed. Selenof antibodies enriched 44 proteins belonging to “structural molecule activity” (marked with red bubbles). Four protein clusters were identified and visualized: “cellular lipid catabolic process” (circled in blue), “Cytosolic ribosome” (circled in yellow), “Intermediate filament” (circled in green), and Selenof along with UGGT1 (C). UGGT1 antibodies enriched 63 proteins belonging to “extracellular exosome” (marked with red bubbles). A cluster with a function “Poly(A) RNA binding proteins” is circled in green in (D). Disconnected nodes are not shown.