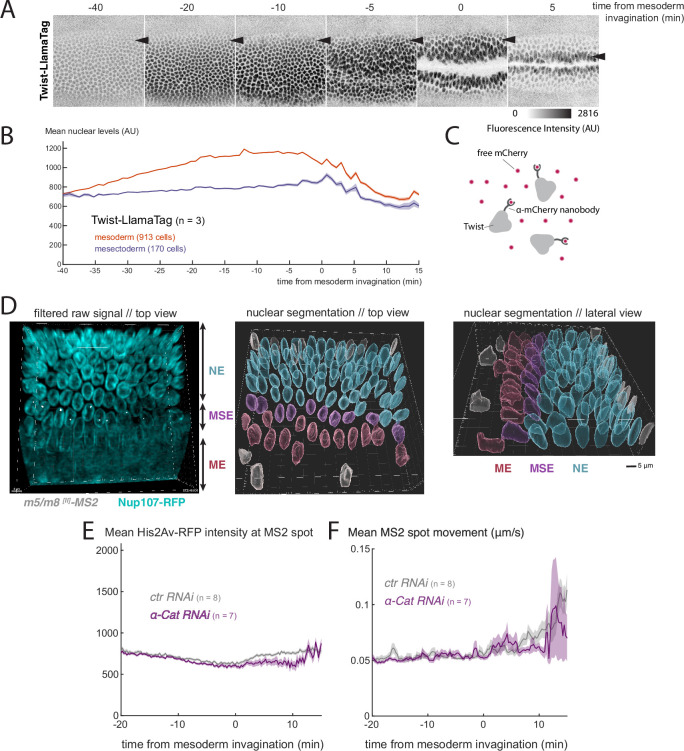
Figure 6. Nuclear levels of Twist and chromatin properties during gastrulation.
(A) Still images from a video of an embryo expressing endogenous Twist fused to LlamaTag, which binds mCherry and allows to visualize nuclear levels of zygotically produced Twist (see C). Arrowhead indicates the position of the mesectoderm, note that Twist levels remain lower in these nuclei than in the central mesoderm. (B) Quantification of nuclear levels of Twist in mesoderm and mesectoderm during gastrulation. (C) LlamaTag approach: Twist is fused to an anti-mCherry nanobody (LlamaTag) that binds free and already fluorescent mCherry that has been maternally loaded in the embryo. (D) Still of the start of mesoderm invagination from a video of an embryo expressing the nuclear membrane marker Nup107-RFP (cyan) and MCP-GFP (grey) to quantify m5/m8 [II] transcription and mark mesectoderm nuclei (left) and segmented nuclei color-coded based on the different cell populations defined from MS2 signal (ME: mesoderm, MSE: mesectoderm, NE: neuroectoderm). Center panel shows view from the top and right panel shows lateral view. (E) Quantification of average His2Av-RFP intensity around the MS2 spot, as a proxy of chromatin compaction, over time in control and α-Cat RNAi embryos. (F) Quantification of the average MS2 spot movement (distance moved relative to the nucleus centroid / time) over time in control and α-Cat RNAi embryos. B, E and F show mean and SEM (shaded area) from all cells combined from multiple embryos (n embryo numbers indicated in each). In F, values were smoothed over time using a median filter of 8 frames (2 min).