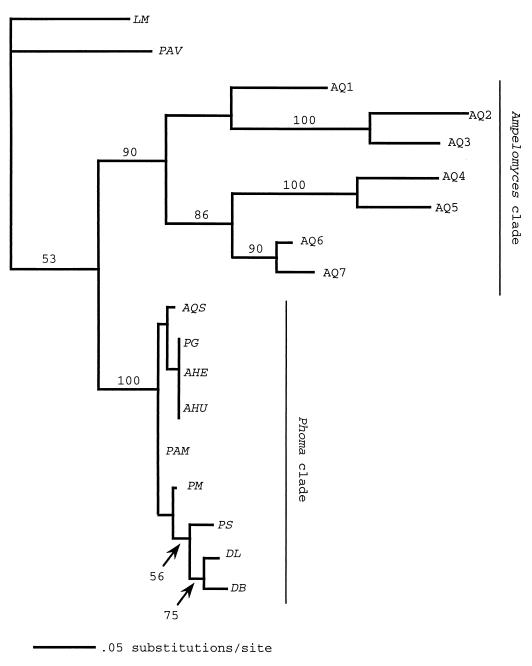
FIG. 2.
Maximum likelihood phylogram based on ITS region sequences between the 18S and 5.8S ribosomal DNA (ITS1). Bootstrap confidence levels (percent) of branches are shown. The scale bar is based on a total tree length of 204. Taxa included in the analysis, GenBank numbers (if known), and their sources are as follows: LM, L. microscopica (LMU04234); PAV, P. avenaria (PAU77357); AQ1, A. quisqualis (AQU82451, DSM [Deutsche Sammlung von Mikrooganismen und Zellkulturen GmbH, Braunschweig, Germany] 2223); AQ2, A. quisqualis (AF126818, South River); AQ3, A. quisqualis (AF126817, ATCC 200245); AQ4, A. quisqualis (AF035782); AQ5, A. quisqualis (AQU82449, CBS [Centraalbureau voor Schimmelcultures, Baarn, The Netherlands] 130.79); AQ6, A. quisqualis (AF035783, Ecogen AQ10); AQ7, A. quisqualis (AF035781, CBS 131.31); AQS, A. quercinus (AF035778, ATCC 36786); PG, P. glomerata (AF126816, South River); AHE, A. heraclei (AF126819, ATCC 36804); AHU, A. humuli (AF035779, ATCC 38616); PAM, P. americana Morgan-Jones et White (AF046016); PM, P. macrostoma Mont. (AF046020); PS, P. sorghina (Sacc.) Boerema (AF046022); DL, D. lycopersici Klebahn (AF046015); and DB, D. bryoniae (Auersw.) Rehm (AF046014).