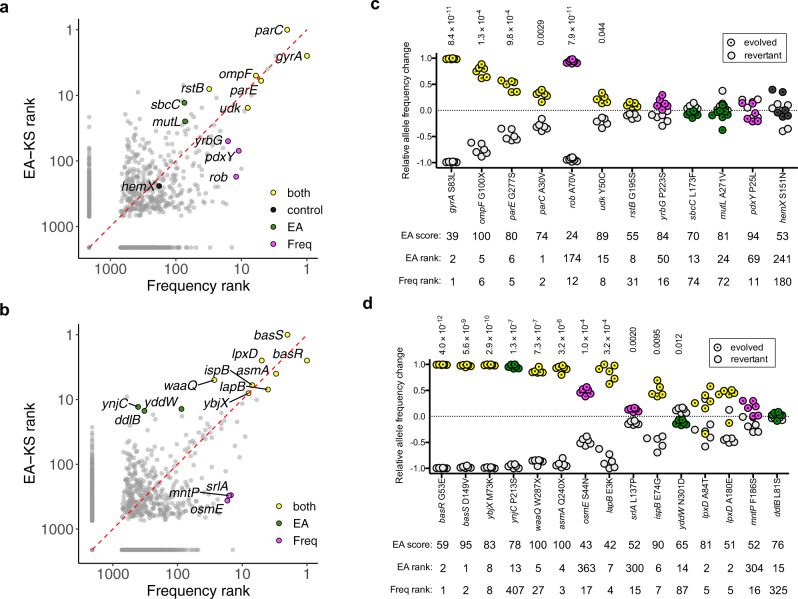
Fig. 3. Mutations in genes highly ranked by both EA integration (EA-KS approach) and frequency analysis contribute to fitness.
a, b The combined rankings for ciprofloxacin and colistin datasets using both EA integration (EA-KS) and frequency-based analysis. Genes that were chosen for subsequent experiments are labeled with their names and colored yellow if ranked highly by both EA integrals and frequency analysis, green if EA specific, and magenta if frequency specific. The evolved strains and isogenic revertants of indicated mutations were competed in the presence of (c) ciprofloxacin or (d) colistin. In the competition assays (c, d), n = 4 independent biological samples for hemX. N = 5 independent biological samples for udk, rstB, and ispB. N = 8 independent biological samples for rob and mutL. All other genes were repeated with n = 6 independent biological samples. Relative allele frequency change after the competition of the evolved allele and the wild type allele are plotted. EA scores (0-100, wherein 100 is highest predicted impact) for each tested mutation are indicated along with the overall rank of each gene based on either EA-KS aggregate ranks or frequency-based aggregate ranks. One sample two-tailed t-test (µ0 = 0) with Bonferroni correction was performed for each competition assay (p values <0.05 reported above each dataset). Source data are provided as a Source Data file.