Figure 1: Workflow of JUMPn.
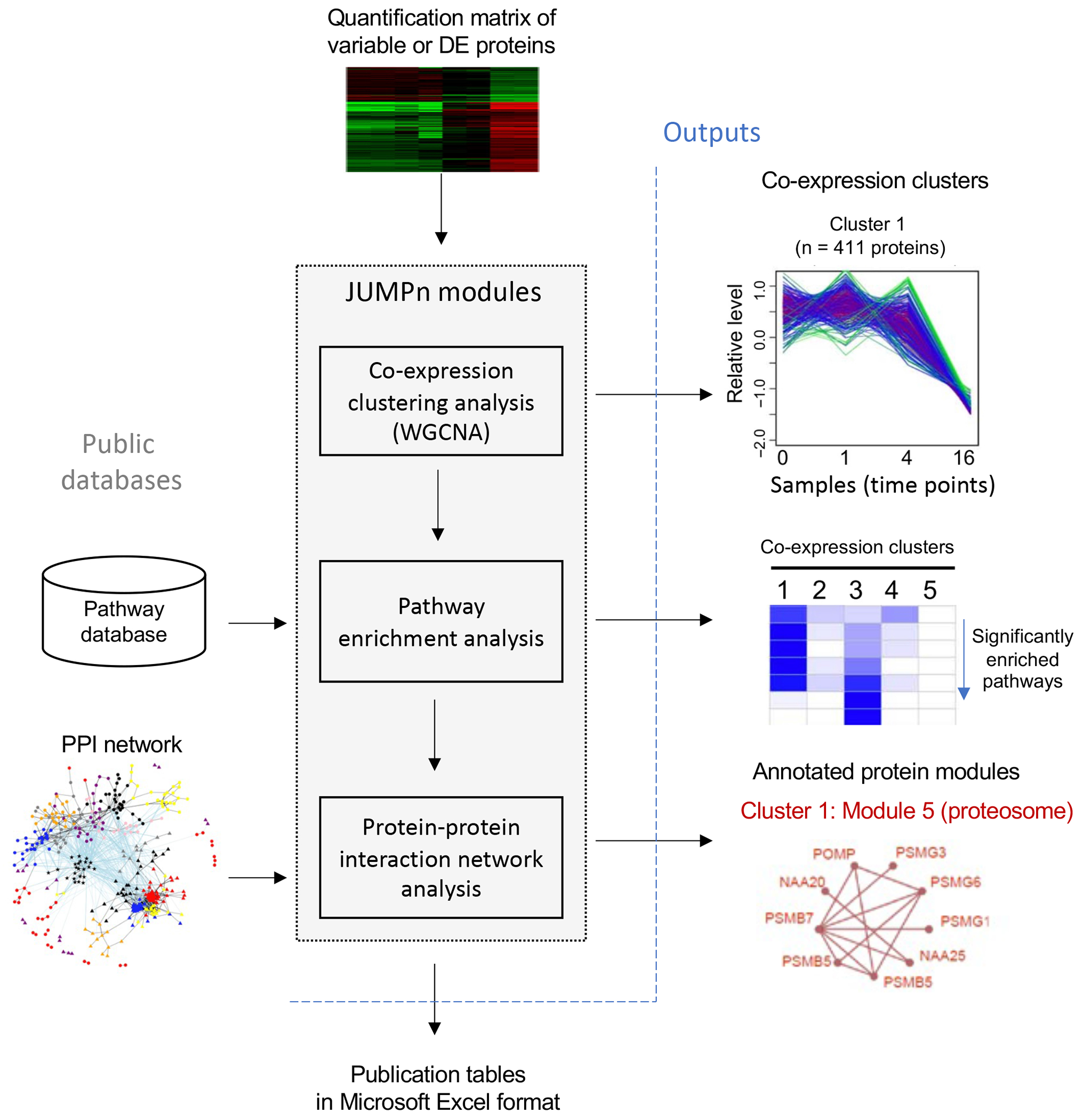
Quantification matrix of the top variable of differentially expressed (DE) proteins are taken as input, and proteins are grouped into co-expression clusters by the WGCNA algorithm. Each co-expression is then annotated by pathway enrichment analysis and further superimposed onto the protein-protein interaction (PPI) network for densely connected protein module identifications.
