Extended Data Fig. 2. Performance of Orca model predictions for cross-cell-type genome interaction difference.
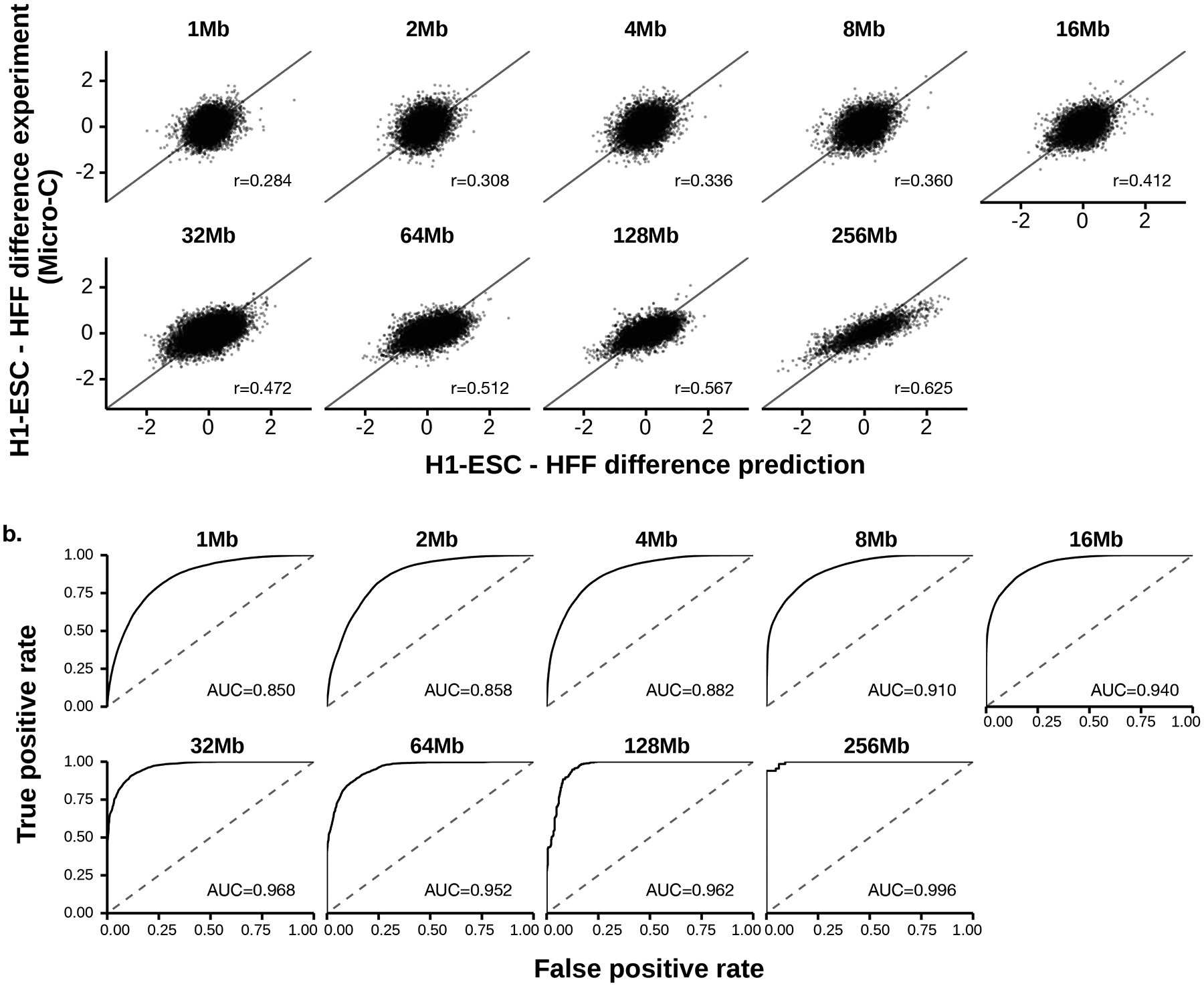
a). Scatter plot comparison of the predicted cell type differences of genome interactions (HFF - H1-ESC) with the micro-C measured interaction score differences on the holdout chromosomes. 10,000 randomly subsampled scores are shown in each panel. The overall Pearson correlations across the entire test chromosomes are annotated. The genome interactions are represented by the log fold over genomic-distance-based background scores for both prediction and experimental data. b). Prediction performance for position pairs with the strongest absolute log-fold differences between the two cell types (top 1 percentile). The performance of models predicting the cell type labels (the cell type with stronger interaction) is measured by receiver operating characteristic (ROC) curve. The area under the ROC curve (AUROC) is annotated. The AUROC score can be interpreted as the probability of a randomly selected positive example (i.e. stronger in HFF) being ranked higher than a randomly selected example (i.e. stronger in H1-ESC). Predictions for 1–32Mb levels are from the Orca-32Mb models and 64–256Mb levels are from the Orca-256Mb models.
