Abstract
Background
Tyrosine kinase inhibitor (TKI) treatment has significantly improved the prognosis of oncogenic-driven lung adenocarcinoma (LUAD). However, drug resistance limits the long-term benefits of patients. Therefore, there is a pressing need to explore the mechanism of TKI resistance and identify new therapeutic targets. It is possible to overcome TKI resistance by inducing tumor cell death through a new process called ferroptosis. Aberrations in ferroptosis, which is a kind of regulated cell death (RCD), has been confirmed to be involved in the development and progression of multiple tumors, and is closely related to patient survival. At present, the role of ferroptosis in TKI resistance remains unclear.
Methods
Ferroptosis-related factors were isolated by expression characteristics analysis based on the multi-omics data of LUADs and normal lung tissues from The Cancer Genome Atlas (TCGA) database. Next, expression of selected ferroptosis-related factors and prognosis were analyzed. Subsequently, the differences in the expression of selected ferroptosis-related factors before and after TKI resistance on a variety of LUAD cell lines were analyzed to identify the factors that were involved in TKI resistance. Finally, the therapeutic effects were confirmed in vitro by targeting the selected ferroptosis-related factors with small molecule compounds.
Results
Glutathione Peroxidase 4 (GPX4), a ferroptosis-related factor, was up-regulated in tumor tissue of LUADs, and correlated with the prognosis of patients. By detecting the expression change of GPX4 before and after TKI resistance in a variety of LUAD cell lines, we confirmed that the inhibition of GPX4 could overcome epidermal growth factor receptor (EGFR)-TKI resistance by inducing ferroptosis.
Conclusions
GPX4 could serve as a novel therapeutic target for EGFR-TKI resistance in LUAD.
Keywords: Ferroptosis, lung adenocarcinoma (LUAD), TKI resistance, GPX4, target
Introduction
Lung cancer is a malignant tumor with high morbidity and mortality rates. Epidemiological statistics have shown that in 2020, the incidence of lung cancer accounted for 11.4% of all malignant tumors, and the mortality rate accounted for 18% of all cancer deaths in the world (1). The major pathological subtypes of lung cancer include squamous carcinoma (LUSC), adenocarcinoma (LUAD), small cell carcinoma (SCLC), and large cell carcinoma (LCLC) (2). It is well known that LUAD has replaced LUSC as the main pathological type of lung cancer, especially in East Asia (3,4). In-depth studies on the pathological behaviors and molecular mechanisms of LUAD have led to the development of systematic clinical treatment strategies for LUAD patients, including chemotherapy, targeted therapy, and immunotherapy (5,6). Of these, targeted therapy is considered to be the most optimal treatment for oncogenic-driven LUAD, owing to its therapeutic specificity and efficacy, which is usually achieved by the application of tyrosine kinase inhibitors (TKIs) (7-9). Although TKI treatment has significantly improved the prognosis of LUAD, the prevalence of drug resistance still limits its long-term benefits to patients. Therefore, there is a pressing need to understand the mechanism of drug resistance towards TKIs and to discover new therapeutic targets (10,11).
Proliferation is the key character of progressive tumors, and thus, overcoming drug resistance towards TKI treatment is achieved via inhibition of tumor cell proliferation or the novel induction of tumor cell death (6,12). The main processes of regulated cell death (RCD) include apoptosis, necroptosis, pyroptosis, and ferroptosis (13-15). Apoptosis is the most extensively studied modality of RCD, but there are currently no effective drugs that can induce the apoptosis of tumor cells (16,17). Ferroptosis is a new mode of RCD, which is caused by the accumulation of iron-dependent lipid peroxidation (18-20). Studies have confirmed that aberrations in ferroptosis are involved in the progression of a variety of malignant tumors, and is also closely related to patient survival (21-23). Previous research have revealed that tumor cells who develop resistance to TKI or cytotoxic drugs via mesenchymal transformation or de-differentiation exhibit high sensitivity to ferroptosis inducers (FINs) (24-27). In addition, the rapid proliferation of tumor cells during disease progression is reflected by active metabolism, high reactive oxygen species (ROS) load, and vigorous iron supply (19,28-30). These characteristics are highly consistent with ferroptosis, which also implies the feasibility of inducing ferroptosis of tumor cells as a treatment strategy after TKI resistance (20,27,31,32). However, targeting ferroptosis as a new therapeutic strategy for LUAD patients with TKI resistance has not yet been elucidated.
In this study, we first describe the expression characteristics of ferroptosis-related factors in LUAD based on multi-omics data from The Cancer Genome Atlas (TCGA) database, and identify several ferroptosis-related factors that were differentially expressed between LUADs and normal lung tissues, among which GPX4 was up-regulated in tumor tissues and significantly correlated with the prognosis of LUAD. We then identify the expression change of GPX4 before and after TKI resistance in a variety of LUAD cell lines, and confirm that inhibition of GPX4 would induce cell ferroptosis, thereby overcoming EGFR-TKI resistance.
In summary, this work systematically exhibited the expression characteristics of ferroptosis-related factors in LUAD as well as their relationship with the prognosis of LUAD patients, in addition to offering a new therapeutic target for EGFR-TKI resistance in LUAD. We present the following article in accordance with the MDAR reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-318/rc).
Methods
Bioinformatics analysis
Ribonucleic acid (RNA) sequencing, whole genome sequencing, deoxyribonucleic acid (DNA) methylation sequencing, and LUAD sample clinical data were isolated from the TCGA database (https://cancergenome.nih.gov/). The Fragments Per Kilobase Million (FPKM) expression data were converted into Transcripts Per Million (TPM) data by using R language (R Foundation for Statistical Computing, version 3.6, Vienna, Austria) to obtain the expression matrix of ferroptosis-related regulatory factors in LUAD. Subsequently, the Wilcox rank sum test was used to analyze the differentially-expressed ferroptosis-related factors between LUADs and normal lung tissues (P<0.05). Clustering heat maps of differentially-expressed ferroptosis-related factors were constructed by using the PHEATMAP software package in R language. Gene ontology (GO) enrichment analysis of differentially-expressed ferroptosis-related factors was performed by using the Cluster profiler software package in R language. The Genvisr software package in R language was used to visualize the mutation and copy number variation (CNV) data of the selected genes. DNA methylation sites in the promoter region were used in the methylation analysis, and the promoter region was defined as 1kb upstream of the gene. For the average value of multiple DNA modification sites in the promoter region, the Wilcox rank sum test was used to analyze ferroptosis-related factors with significant differences in methylation between LUAD and normal lung tissue (P<0.05).
Biological tissue samples
From September 2020 to January 2021, 12 patients with lung adenocarcinoma (ADC) and their matched adjacent non-tumor tissues were collected from the Department of Thoracic Surgery, West China Hospital, Sichuan University. A part of it was temporarily stored at −80 ℃ to extract total RNA and total protein, and real-time fluorescence quantitative PCR (RT-qPCR) and Western blot experiments were carried out respectively; The rest was fixed with 10% neutral formalin and embedded in paraffin for immunohistochemistry (IHC) analysis.
Tissue microarrays (HLugA180Su04) containing 82 pairs of lung adenocarcinoma patients and their matched adjacent lung tumor tissues were purchased from Shanghai Outdo Biotech Co., Ltd. (China). Clinicopathological data included patients’ age, gender, tumor size, histological type, pathological grade, tumor lymph node metastasis (TNM) stage, lymph node metastasis status, survival status and overall survival (OS) time. OS was defined as the time from treatment to death or the last follow-up. Tissue microarrays were used for immunohistochemical experiments to evaluate the correlation between GPX4 expression and the prognosis of LUAD patients.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Institutional Review Board of West China Hospital, Sichuan University (No. 2016-98) and informed consent was taken from all the patients.
Real-time quantitative fluorescence PCR
Total RNA from clinical samples and cell lines used in this study were extracted by using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The Complementary Deoxyribonucleic Acid (cDNA) synthesis was performed with PrimeScript™ Reverse Transcriptase kit (Takara, Dalian, China). Next, the mRNA expression levels of GPX4, NFE2 Like BZIP Transcription Factor 2 (NRF2), Kelch Like Epichlorohydrin Associated Protein 1 (KEAP1), Ferritin Heavy Chain 1 (FTH1), Acyl-CoA Synthetase Long Chain Family Member 4 (ACSL4), and Lysophosphatidylcholine Acyltransferase 3 (LPCAT3) were qualified by real-time quantitative polymerase chain reaction (RT-qPCR) by using the SYBR PremixEx Taq TM II kit (Takara, RR820A, Dalian, China). The signal was detected by using a Real-time fluorescence quantitative PCR instrument (Bio-Rad, CFX Connect, Hercules, CA, USA). The human 18 Svedberg Ribosomal RNA (18S rRNA) was used as the endogenous control for quantifying the expression of selected genes. The relative gene expression levels were calculated by using the comparative 2−DeltaDelta Cycle threshold (2−ΔΔCt) method.
The real-time PCR primers were as follows: Human GPX4 (forward primer: 5'-CAGTTCGGGAAGCAGGAG-3'; reverse primer: 5'-GCCCTTGGGTTGGATCTT-3'); Human NRF2 (forward primer: 5'-ATATTCCCGGTCACATCG-3'; reverse primer: 5'-CCAAACTTGCTCAATGTCC-3'); Human KEAP1 (forward primer: 5'-AGAAGTCCAAAGCGGGAA-3'; reverse primer: 5'-CTATTACCCGGCCAGAGG-3'); Human FTH1 (forward primer: 5'-AGTCGTCGGGGTTTCCT-3'; reverse primer: 5'-GAGGGTGCGGTGAAGAG -3'); Human ACSL4 (forward primer: 5'-CATACACATCAGCCATCAACT-3'; reverse primer: 5'-TATCAAGGAGGCAGGAACA-3'); Human LPCAT3 (forward primer: 5'-CTTGTGGGAGGAGCAGAG-3'; reverse primer: 5'-GTGGTAGCGGCACATACA-3'); Human 18S rRNA (forward primer: 5'-GATGGGCGGCGGAAAATAG-3'; reverse primer: 5'-GCGTGGATTCTGCATAATGGT-3').
Western blotting
The fresh-frozen tissues and whole-cell lysates were extracted by using radioimmunoprecipitation assay (RIPA) buffer (high) (R0010, Solarbio, Beijing, China) with a protease inhibitor mixture (P6730, Solarbio, Beijing, China). The tissue and cell lysates were centrifuged at 14,000 rpm for 15 min at 4 ℃. Aliquots of supernatant protein concentration was measured by using a Bicinchoninic acid (BCA) Protein Assay Kit (PC0020, Solarbio, Beijing, China), and then the protein lysate was mixed with 5× loading buffer and denatured at 100 ℃ for 5 min. An equal amount of proteins (30 µg) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and electrophoretically transferred to a polyvinylidene fluoride (PVDF) membrane (Merck Millipore, Cork, IRL). The PVDF membranes were then treated for 1 hour by using 5% (w/v) non-fat milk at room temperature. After incubating with anti-GPX4 (1:1,000; CST, #52455), anti-NRF2 (1:1,000; CST, #12721), anti- KEAP1 (1:1,000; CST, #8047), anti-FTH1 (1:1,000; CST, #4393), anti-ACSL4 (1:500; Santa Cruz, sc-365230), and anti-LPCAT3 (1:1,000; Creative Biolab, CBMAB-L0337-YC) overnight at 4 ℃, the PVDF membranes were washed with 1× Tris Buffered Saline with Tween-20 [TBST, TBS + 0.1% (v/v) Tween-20] and incubated with corresponding horseradish peroxidase (HRP)-conjugated secondary antibody (1:5,000; Invitrogen) at room temperature. Then, the immunoreactive bands were visualized via using enhanced chemiluminescence (ECL) detection reagent (Beijing 4A Biotech Co., Ltd.). The expression of β-actin (1:2,000; ZSGB-BIO, TA-09) was used as the loading control.
Immunohistochemistry and quantification analysis
Surgically derived tissues were fixed in 10% phosphate-buffered formalin by using a graded series of ethanol, embedded in paraffin, sectioned at a thickness of 4 µm, and finally mounted on glass slides. The tissue sections were roasted at 65 ℃ for 4 hours, and then de-paraffinized and rehydrated with xylene and degraded alcohol. The slices were immersed in 1× citrate repair solution and heated in microwave oven until boiling for 16 min. The slices were cooled to room temperature on the laboratory bench. After washing with 0.01 M phosphate buffered saline (PBS) buffer, 3% hydrogen peroxide (H2O2; v/v) was used for 10 min to block endogenous peroxidase activity and to avoid immune reaction. Then, the sections were blocked with normal goat serum for 10 min and then incubated with the GPX4 primary antibody (1:200; ZEN BIO, 381958) at 4 ℃ overnight. After washing with PBS, the sections were incubated with HRP-conjugated anti-rabbit antibody (PV-6000; ZSGB-BIO) at room temperature for 1 hour, stained with 3'3-diaminobenzidine (DAB) substrate solution (ZLI-9018, ZSGB-BIO) at room temperature for an appropriate time, and then counterstained with hematoxylin for 1–5 min.
Two researchers blindly and independently assessed the stained slides. According to the semi-quantitative scoring system, the GPX4 protein expression level was calculated along the staining intensity and the percentage of positive cell. The staining intensity was scored 0 for negative, 1 for weak, 2 for moderate, and 3 for strong. A proportion of positive cells ≤5% was scored 0, 6–25% was scored 1, 26–50% was scored 2, 51–75% was scored 3, and 76–100% was scored 4. The immunoreactivity score was obtained by multiplying these two scores (33). Patients were divided into two groups according to the GPX4 immune response score: immunoreactivity scores <6 were placed into the low GPX4 expression subgroup, while immunoreactivity scores ≥6 were placed into the high GPX4 expression subgroup.
Cell culture and reagents
Human bronchial epithelial cell lines (HBE, MRC-5, and BEAS-2B) were kindly provided by Prof. Peng Yong in 2016 (State Key Laboratory of Biotherapy, West China Hospital, Sichuan University). Human lung cancer cell lines (A549, H292, H446, and H460) were provided by the Core Facility of West China Hospital, Sichuan University. Several other lung cancer cell lines, including H1975 (EGFR mutation: L858R/T790M) (CBP60121), H1299 (CBP60053), HCC78 (ROS1 fusion: SLC34A2-ROS1) (CBP60100), HCC827 (EGFR mutation: delE746-A750) (CBP60101, PC-9 (EGFR/del E746-A750) (CBP73212), and H3122 (ALK fusion: EML4-ALK E13; A20) (CBP60133), were purchased from Nanjing Cobioer Biotechnology Co., Ltd. (China). Several drug-resistant cells (HCC78CR, HCC827GR, H3122LR, PC9OR) used in this study were constructed in our laboratory.
BEAS-2B (B2B) was cultured in BEGM™ Bronchial Epithelial Cell Growth Medium (BEGM medium, Lonza, USA); A549 was cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Waltham, MA, USA), and the remaining cells were cultured in Roswell Park Memorial Institute 1640(RPMI1640 medium, Gibco, Waltham, MA, USA). All cell lines were cultured at 37 ℃ in a humidified atmosphere of 5% carbon dioxide (CO2), and culture media (except BEGM) were added 10% fetal bovine serum (FBS, Lonsera, Uruguay, South America), 1% antibiotics (Invitrogen, Carlsbad, CA, USA; Penicillin 100 U/mL and streptomycin 100 mg/mL), and Plasmocure Anti-mycoplasma agent (ant-pc, Invivogen, USA). The cell lines were authenticated by short tandem repeats (STR) profiling and confirmed to be free of mycoplasma contamination.
Cell viability assay
TKI sensitive and resistant cell lines in the exponential phase were digested, centrifuged, and resuspended into a single cell suspension. The cells were seeded in 96-well plates at a density of 5,000 cells/well, respectively, and cultured overnight to ensure cell adherence. They were then treated with different concentrations (1S, 3R)-2-(2-Chloroacetyl)-2,3,4,9-tetrahydro-1-(4-(methoxycarbonyl)phenyl)-1H-pyrido(3,4-b)indole-3-carboxylic acid methyl ester (RSL3, 0–20 µmol/L, HY-100218A, MedChemExpress, USA) and Erastin (0–20 µmol/L, HY-15763, MedChemExpress, USA) for another 72 h, respectively, to determine the half maximal inhibitory concentration (IC50) values of RSL3 and Erastin in each cell line. To determine the ability of RSL3 to overcome TKI resistance, RSL3 was used at the concentration corresponding to the IC25 value of each cell line.
For PC9OR, the addition concentration of RSL3 was 180 nM, and that of osimertinib (HY-15772S, MedChemExpress, USA) were 100, 500, 1,000, 2,000, 4,000, 8,000, 16,000, and 20,000 nM. For HCC78-CR, the addition concentration of RSL3 was 20 nM, and that of crizotinib (S1068, Selleck Chemicals, Shanghai, China) were 10, 100, 500, 1,000, 5,000, 10,000, 20,000, and 30,000 nM. For H3122LR, the addition concentration of RSL3 was 135 nM, and that of lorlatinib (S7536, Selleck Chemicals, Shanghai, China) were 10, 50, 100, 250, 500, 1,000, 5,000, 10,000, 20,000, and 30,000 nM. For HCC827GR, the addition concentration of RSL3 was 50nM, and that of gefitinib (HY-50895, MedChemExpress, USA) were 10, 100, 500, 1,000, 5,000, 10,000, 20,000, 30,000, 40,000, and 50,000 nM. Three parallel control wells were set for each concentration.
Following treatment, fresh medium containing 10% cell counting kit-8 (CCK-8, HY-K0301, MedChemExpress, USA) reagent (100 µL/well) was added and incubated at 37 ℃ for 1h. A gene microplate reader (Fludia T70; Photonics Technology International, New Jersey, USA) was used to detect the absorbance (OD) at a wavelength of 450 nm. Cell viability was calculated by dividing the absorbance change of the treated cells by the percentage of absorbance of the untreated cells. We then plotted the growth inhibition curve and calculated the IC50.
ROS determination
ROS production was detected by using 6-carboxy-2',7'-dichlorofluorescein diacetate dye (DCFH-DA; CA1410; Beijing Solarbio Science & Technology Co., Ltd) (34). DCFH-DA was diluted with serum-free medium at 1:1,000 to a final concentration of 10 µmol/L, and the medium containing DCFH-DA was replaced with cells treated with RSL3, osimertinib, gefitinib, or Dimethyl sulfoxide (DMSO). The cells were incubated in an incubator at 37 ℃ for 20 min, washed with serum-free cell culture solution three times to fully remove the non-intracellular DCFH-DA, and then digested with trypsin. ROS production was detected by using a flow cytometer (Cytoflex, Beckman Coulter, USA) analyzed with CytExpert software (Beckman Coulter, USA).
Propidium iodide (PI) staining
TKI sensitive and resistant cells were seeded in 12-well plates, and treated with RSL3 (1 µM), osimertinib (1 µM), gefitinib (1 µM), or DMSO for another 8–24 h after 60–80% adherent fusion. The cells were harvested and washed twice with PBS, then re‑suspended in a PI (50 µg/mL) staining solution at room temperature for 5 min in the dark. After adding 200 µL of chilled PBS and gently mixing, the prepared cells were detected by using a Beckman Cytoflex flow cytometer (Beckman Coulter, USA). At least 20,000 events were collected and the data were analyzed by using CytExpert software (Beckman Coulter, USA).
Immunofluorescence
The cells were treated in a formalin solution (10%; v/v) for 15 min and permeabilized with Triton X-100 for 10 min at room temperature. They were then blocked with normal goat serum for 10 min and then incubated with the primary antibody (GPX4, 1:200, Proteintech, Cat No. 67763-1-Ig) at 4 ℃ overnight. After washing with PBS three times, the cells were incubated with fluorescein-conjugated secondary antibody (Alexa Fluor-488, Goat Anti-Mouse) in the dark for 1 hour at room temperature, and then stained with 4,6-diamidino-2-phenylindole dihydrochloride for 5 min. After washing three times with PBS, and the stained cells were mounted by using an anti-fade mounting medium (S2100, SolarBio, Beijing, China), covered with coverslips, and images were captured by using an epifluorescence microscope (Nikon, Tokyo, Japan).
Statistical analysis
The statistical methods involved in the bioinformatics analysis were performed as mentioned above. The data were expressed as mean ± standard deviation (SD). Statistical analysis and mapping were performed by using R language (version 3.6), GraphPad Prism 8 (GraphPad Inc., La Jolla, CA, USA), and SPSS 22.0 (SPSS Inc., Chicago, IL, USA) software. The expression differences in GPX4 between LUAD and matched non-tumor normal tissues were compared by using the t-test, and the other statistical analyses were executed by employing the Wilcox rank sum test. The relationship between GPX4 and the clinicopathological variables was analyzed via the Pearson chi-square test. The survival curve was plotted by using the Kaplan-Meier method, and significant differences were evaluated by the log-rank test. The expression error bar (RT-qPCR) of target gene in cell lines and tissue are presented as means ± SD (n=3 independent repeats). P values were isolated via by using two-tailed unpaired Student’s t-tests, and P<0.05 was considered statistically significant.
Results
The expression of ferroptosis-related factors in LUAD
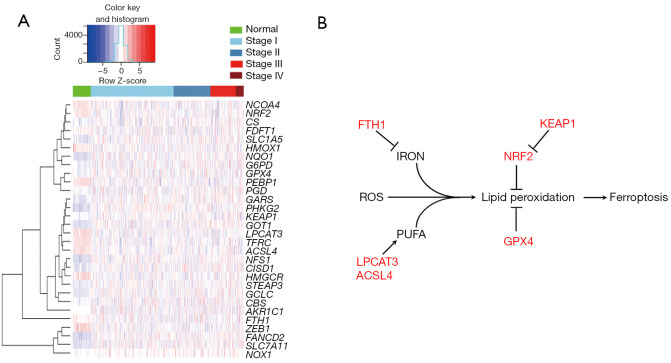
Ferroptosis-related factors were identified by using the Kyoto Encyclopedia of Genes and Genomes (KEGG) ferroptosis signaling pathway, and a total of 42 ferroptosis-related factors were screened (Table S1). Based on the TCGA LUAD transcriptome data analysis, a total of 30 ferroptosis-related factors were found to be expressed differently between LUADs and normal lung tissues. The expression of ferroptosis-related factors was significantly different in LUADs compared to normal lung tissues, which indicated the different ferroptosis sensitivities of tumors among LUAD patients (Figure 1A).
Figure 1.
Differentially-expressed ferroptosis-related factors between LUAD and normal lung tissues. (A) The heatmap of differentially-expressed ferroptosis-related factors between LUAD and normal lung tissues based on TCGA LUAD transcriptome data. (B) Selected six key ferroptosis regulators with differential expression between LUAD and normal lung tissues, and their role on ferroptosis. LUAD, lung adenocarcinoma; TCGA, The Cancer Genome Atlas.
Genomically, a total of 38 ferroptosis-related factors had undergone CNV; CNV occurred in 357 samples, with a CNV amplification rate of 0.21–10.71%, accounting for 64.99% (232/357) (Figure S1A; Table S2). Also, 31 ferroptosis-related factors were found to have gene mutations in 476 samples (Figure S1B). DNA methylation analysis showed that 16 ferroptosis-related factors exhibited significantly different DNA methylation levels on the promoter region between LUADs and normal lung tissues (Figure S1C).
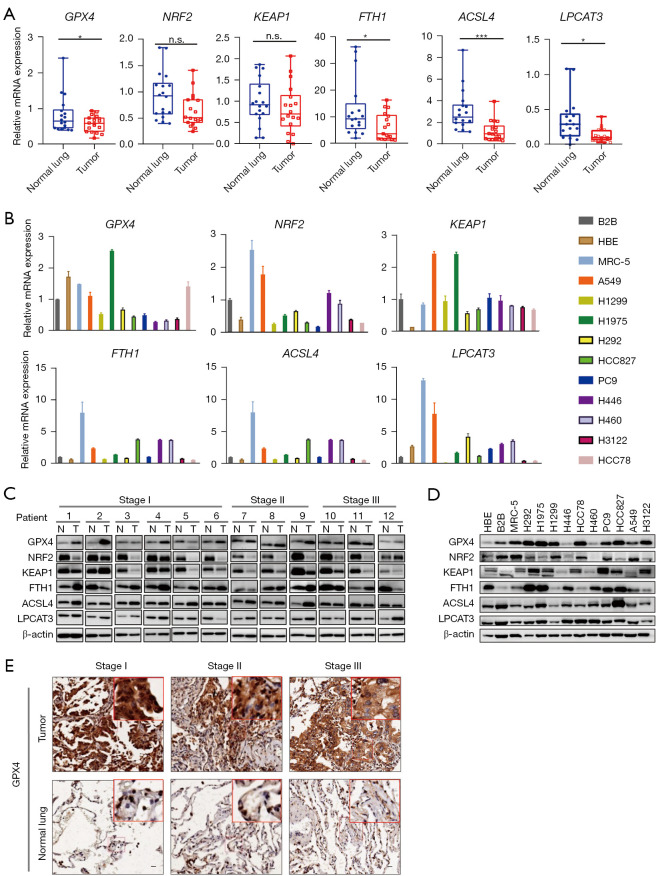
We selected six key regulatory factors of ferroptosis from different expressed genes for our subsequent study, including GPX4, NRF2, KEAP1, FTH1, ACSL4 and LPCAT3. Of these, GPX4, NRF2, KEAP1and FTH1 acted as negative regulators, while ACSL4 and LPCAT3 were positive regulators (Figure 1B). RT-qPCR detection of clinical samples revealed that GPX4, LPCAT3, ACSL4, and FTH1 were expressed at low levels in LUADs compared with normal lung tissues (P<0.05), while there was no significant difference in the mRNA expression of NRF2 and KEAP1 between LUADs and normal lung tissues (Figure 2A). We also detected the mRNA expression of these factors in LUAD cell lines. Compared with normal lung bronchial epithelial cell lines (HBE, B2B, and MRC-5), GPX4 mRNA expression in LUAD cell lines (H1299, H292, H446, H460, HCC78, PC-9, HCC827, A549, H3122) was generally lower, exhibiting a consistent trend with clinical samples. In contrast to GPX4, NRF2, KEAP1, FTH1, ACSL4, and LPCAT3 did not showed consistent expression trend in LUAD cell lines compared to normal lung bronchial epithelial cell lines (Figure 2B). Western blotting showed that GPX4, FTH1, ACSL4, and LPCAT3 were more up-regulated in LUADs than in normal lung tissue, while NRF2 and KEAP1 were down-regulated in LUADs (Figure 2C; Table S3). GPX4 protein expression in LUAD cell lines (H1975, HCC78, PC-9, HCC827, H292, H1299, and H3122) was generally higher, exhibiting a consistent trend with clinical samples. However, NRF2, KEAP1, FTH1, ACSL4 and LPCAT3 showed no obvious expression trend in LUADs cell lines and normal lung bronchial epithelial cell lines (Figure 2D). Immunohistochemistry staining also showed that GPX4 protein expression was up-regulated in LUADs compared with normal lung (Figure 2E). These findings suggested that GPX4 could be the most likely ferroptosis-related factor involved in LUAD progression.
Figure 2.
Expression of selected six key ferroptosis regulators in LUAD. (A) Relative mRNA expression of GPX4, LPCAT3, KEAP1, NRF2, ACSL4, and FTH1 in LUAD and matched normal lung tissues. (B) Relative mRNA expression of GPX4, LPCAT3, KEAP1, NRF2, ACSL4, and FTH1 in LUAD and lung bronchial epithelial cell lines. (C) Protein expression levels of GPX4, LPCAT3, KEAP1, NRF2, ACSL4, and FTH1 in LUAD and matched normal lung tissues. (D) Protein expression levels of GPX4, LPCAT3, KEAP1, NRF2, ACSL4, and FTH1 in LUAD and lung bronchial epithelial cell lines. (E) Representative immunohistochemical images of GPX4 in different stage LUADs and matched normal lung tissues, scale bar =20 µm. *, P<0.05; ***, P<0.001. N, normal tissue; T, tumor tissue; n.s., not significant; LUAD, lung adenocarcinoma; mRNA, messenger RNA.
GPX4 expression significantly correlates with the prognosis of LUAD patients
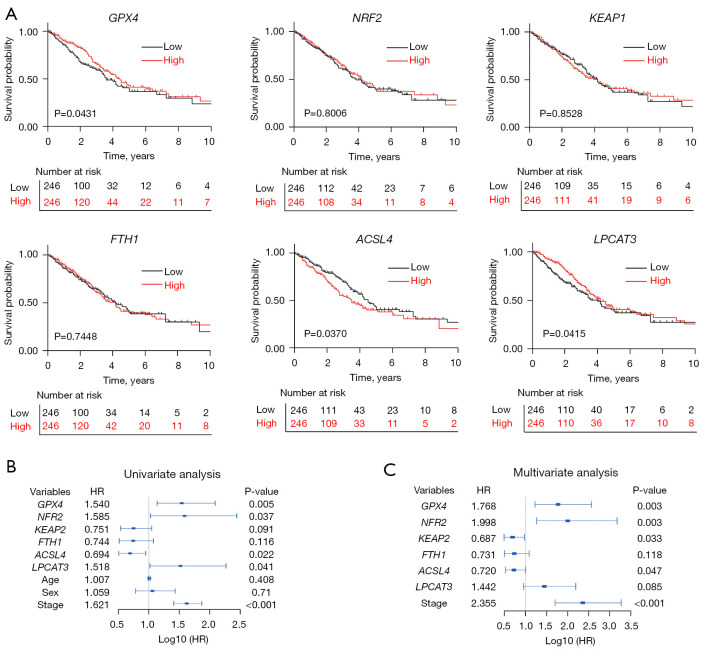
The Kaplan-Meier survival curve based on the TCGA database suggested that GPX4 was associated with the OS of LUAD patients. The prognosis of patients with high GPX4 expression was better than those with low GPX4 expression (median survival time: 53.3 vs. 41 months, P<0.05) (Figure 3A). A Cox proportional hazard regression model was used to evaluate the risk of GPX4 expression on the prognosis of LUAD, which showed that low GPX4 expression was a risk factor for poor prognosis in LUAD patients. Variables with P≤0.1 in the Cox univariate regression analysis were included in Cox multivariate regression model for further analysis, which showed that low GPX4 expression and TNM staging were independent risk factors for lower OS in LUAD patients (Figure 3B,3C; Table S4).
Figure 3.
Correlation between the expression of six key ferroptosis regulators and the survival of LUAD patients based on TCGA database. (A) Kaplan-Meier survival curve of GPX4, LPCAT3, KEAP1, NRF2, ACSL4, and FTH1 in LUAD. Cox regression analysis of GPX4, LPCAT3, KEAP1, NRF2, ACSL4, and FTH1 in LUAD: (B) univariate Cox analysis; (C) multivariate Cox analysis. High, high expression; Low, low expression; HR, hazard ratio; LUAD, lung adenocarcinoma; TCGA, The Cancer Genome Atlas.
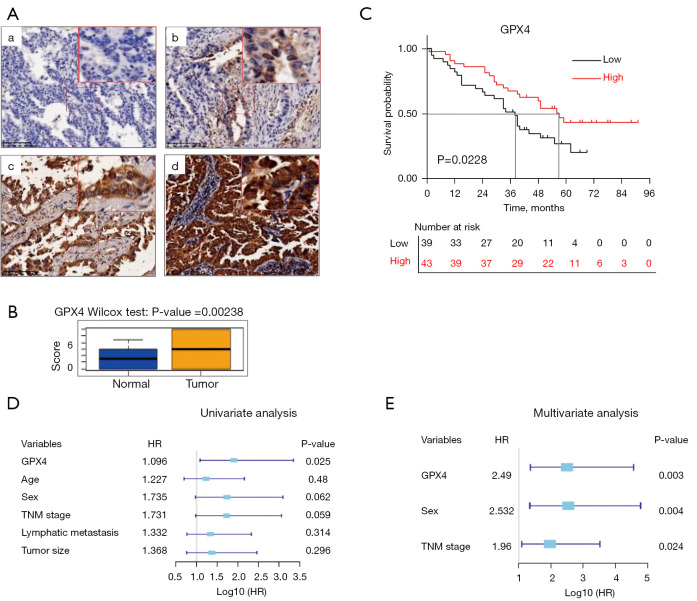
Through immunohistochemical staining of 82 pairs of clinical LUAD samples, we confirmed that there was strong positive expression of GPX4 in LUAD tissue, while its expression in normal alveolar epithelial and bronchial epithelial cells was negative or weakly positive (Figure 4A), and the difference was statistically significant (P=0.00238, Figure 4B). Furthermore, there was no significant correlation between GPX4 expression and age, gender, tumor size, TNM stage, or lymph node metastasis in LUAD patients; GPX4 expression was only related to the histological subtypes of samples (P=0.005, Table S5).
Figure 4.
Correlation between GPX4 expression and the survival of patients based on the LUAD tissue microarray. (A) Representative images of different immunohistochemical staining intensities of GPX4 in the LUAD tissue microarray: (a) negative staining; (b) weak staining; (c) moderate staining; (d) strong staining. Scale bar =200 µm. (B) Different GPX4 expression between LUADs and normal lung tissues. Cox regression analysis of GPX4 in LUADs from the tissue microarray. (C) The Kaplan-Meier survival curve showed that LUAD patients with low GPX4 expression had an unfavorable OS (P=0.0228). (D) Univariate analysis. (E) Multivariate analysis. High, high expression; Low, low expression; HR, hazard ratio.
Survival analysis suggested that the GPX4 expression level was associated with the prognosis of LUAD patients, which was consistent with TCGA data. The median survival time of patients with high GPX4 expression was 57.0 months, which was significantly longer than patients with low GPX4 expression (38.0 months, P=0.0228), and their prognosis was also better (Figure 4C). Cox proportional risk regression analysis suggested that low GPX4 expression was a risk factor for lower OS of LUAD patients (Figure 4D). Age, gender, TNM stage, lymph node metastasis, and tumor size were not associated with OS. Subsequent multivariate Cox proportional risk regression analysis found that low GPX4 expression, male, and advanced TNM were independent risk factors for lower OS in LUAD (Figure 4E, Table 1).
Table 1. Cox regression analysis of the GPX4 protein in LUAD patients from the tissue microarray.
| Variables | Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | ||
| GPX4 expression (low vs. high) | 1.906 | 1.085–3.349 | 0.0248* | 2.490 | 1.359–4.562 | 0.003* | |
| Age (years) (<65 vs. ≥65) | 1.227 | 0.696–2.163 | 0.48 | – | – | – | |
| Sex (male vs. female) | 1.735 | 0.972–3.096 | 0.0622 | 2.532 | 1.340–4.784 | 0.004* | |
| TNM stage (III–IV vs. I–II) | 1.731 | 0.979–3.06 | 0.059 | 1.962 | 1.093–3.520 | 0.024* | |
| LNM (yes vs. no) | 1.332 | 0.763–2.324 | 0.314 | – | – | – | |
| Tumor size (cm) (≤3 vs. >3) | 1.368 | 0.760–2.464 | 0.296 | – | – | – | |
*, P<0.05. LUAD, lung adenocarcinoma; LNM, lymph node metastases; TNM, tumor-node-metastasis; HR, Hazard ratio; CI, confidence interval.
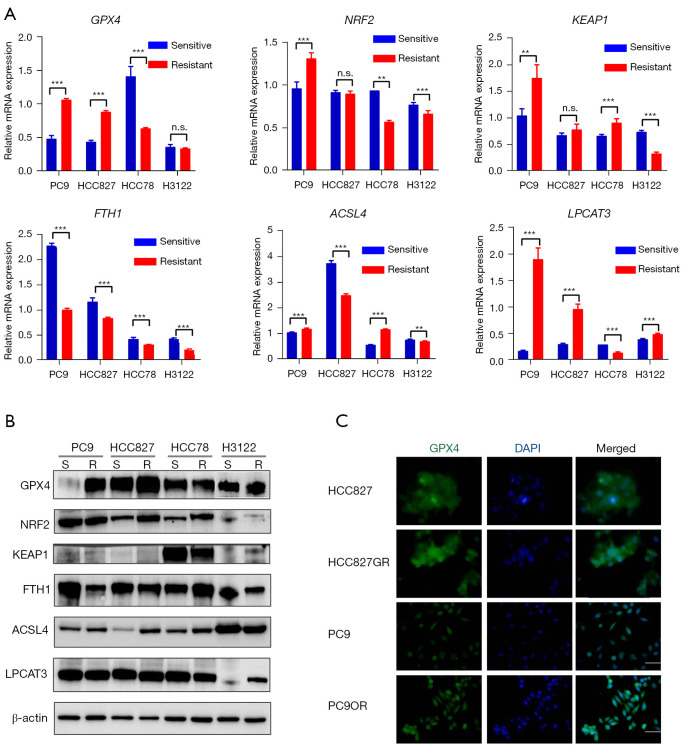
Up-regulation of GPX4 in LUAD cell lines after EGFR-TKI resistance
We then detected the expression of six selected ferroptosis-related factors in TKI sensitive and resistant LUAD cell lines. The mRNA expression of GPX4 was significantly up-regulated in the HCC827GR (gefitinib resistant) and PC9OR (osimertinib resistant) cell lines, in contrast to parental cell lines (HCC827, PC9). Meanwhile, the mRNA expression of GPX4 was significantly down-regulated in HCC78CR (crizotinib resistant) cells in contrast to HCC78 cells, and there was no significant change in GPX4 between H3122 and H3122LR (lorlatinib resistant) cells (Figure 5A). Western blotting and immunofluorescence staining further confirmed the expression changes of GPX4 at the protein level (Figure 5B,5C). However, NRF2, KEAP1, FTH1, ACSL4, and LPCAT3 exhibited no consistent expression change at both the mRNA and protein levels (Figure 5A,5B). These results suggested that GPX4 was especially up-regulated in LUAD cells after EGFR-TKI resistance (HCC827GR, gefitinib; PC9OR, osimertinib), with GPX4 acting as a ferroptosis inhibitor. This also indirectly implies that EGFR-TKI-resistant cells may be sensitive to FINs, targeting GPX4 in particular.
Figure 5.
The expression of GPX4 changed significantly after EGFR-TKI resistance. (A) Relative mRNA expression of GPX4, LPCAT3, KEAP1, NRF2, ACSL4, and FTH1 in TKI-sensitive and TKI-resistant LUAD cell lines. (B) The protein expression levels of GPX4, LPCAT3, KEAP1, NRF2, ACSL4, and FTH1 in TKI-sensitive and TKI-resistant LUAD cell lines. (C) Immunofluorescence revealed that the expression of GPX4 was up-regulated in EGFR-TKI resistant cell lines. Scale bar =20 µm. **, P<0.01; ***, P<0.001. n.s., not significant; EGFR, epidermal growth factor receptor; TKI, tyrosine kinase inhibitor; LUAD, lung adenocarcinoma.
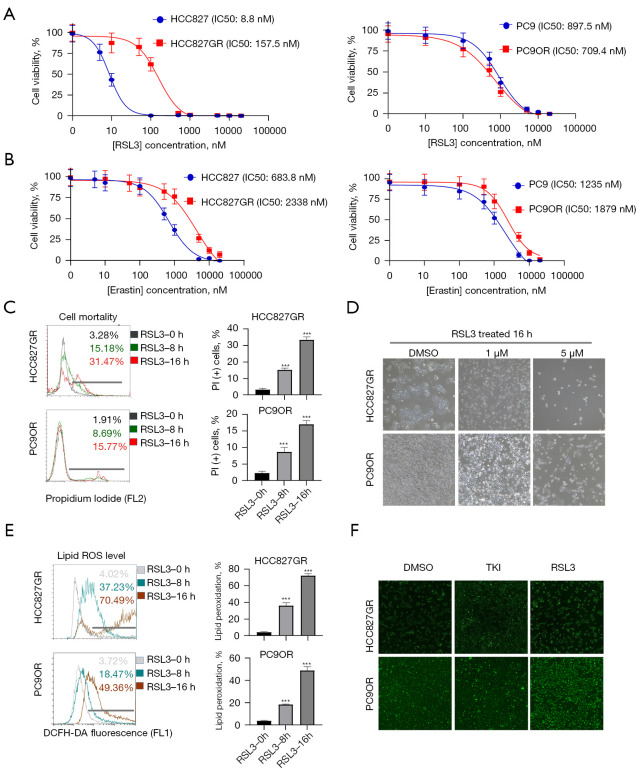
Targeting GPX4 induced the ferroptosis of EGFR-TKI resistant cells
We subsequently treated EGFR-TKI-sensitive (HCC827, PC-9) and EGFR-TKI-resistant (HCC827GR, PC-9OR) cells with RSL3, which is a compound targeting GPX4, to investigate the role of ferroptosis in TKI resistance, and the type I FIN, Erastin, was used as a control (34,35). After treatment for 72 hours, the CCK-8 cell proliferation assay showed that RSL3 and Erastin had dose-dependent killing effects on the above EGFR-TKI-sensitive and EGFR-TKI-resistant cells. The mean IC50 concentration of RSL3 in HCC827 and PC-9 cells was 290±431 nM, and that in HCC827GR and PC-9OR cells was 234±222 nM (Figure 6A), which was significantly lower than that of Erastin (1,871±2,054 and 2,335±37 nM, respectively) (Figure 6B). We verified that the death of EGFR-TKI-resistant cells induced by RSL3 was ferroptosis by observing the morphology and ROS accumulation (Figure 6C-6F). These results implied that GPX4 could serve as a novel therapeutic target for LUAD after EGFR-TKI resistance.
Figure 6.
By targeting GPX4, RSL3 induced EGFR-TKI-resistant LUAD cell death via ferroptosis. RSL3 exerted stronger dose-dependent killing effects on EGFR-TKI resistant cells than Erastin. (A) Proliferation inhibition curves of HCC827GR/HCC827 and PC9OR/PC-9 cells after treatment with RSL3. (B) Proliferation inhibition curves of HCC827GR/HCC827 and PC9OR/PC-9 cells after treatment with Erastin. (C) PI staining analysis of HCC827GR and PC9OR cells after exposure to RSL3 (1 µM) for 0, 8, and 16 h. (D) Representative phase-contrast microscopy images of HCC827GR and PC9OR cells after treatment with 1 and 5 µM RSL3 for 16 h, respectively (magnification, ×100). (E) HCC827GR and PC9OR cells were treated with 1 µM RSL3 for 0, 8 and 16 h, respectively, and then with DCFH-DA for 20 min to detect the ROS level by flow cytometry. (F) Immunofluorescence revealed the aggregation of ROS in HCC827GR and PC9OR cells (magnification, ×100). Data are exhibited as the mean ± standard error based on at least three independent experiments. Significant differences were defined by one-way Analysis of Variance (ANOVA) with a Bonferroni post-hoc test. ***, P<0.001. ROS, reactive oxygen species; DCFDA, 6-carboxy-2',7'-dichlorofluorescein diacetate dye; EGFR, epidermal growth factor receptor; TKI, tyrosine kinase inhibitor; LUAD, lung adenocarcinoma.
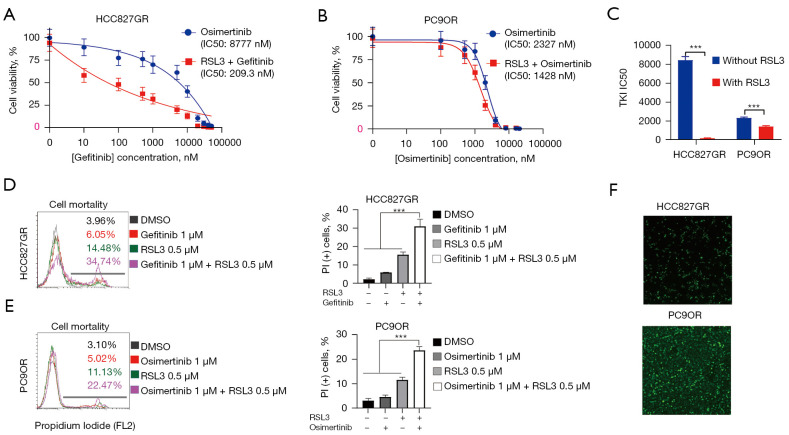
RSL3 enhanced the cytotoxicity of gefitinib/osimertinib in EGFR-TKI-resistant LUAD cells
We then evaluated the therapeutic effect of RSL3 on EGFR-TKI-resistant cells by treating them with RSL3 and EGFR-TKI, and confirmed that RSL3 could enhance the cytotoxicity of gefitinib/osimertinib in HCC827GR and PC-9OR cells, respectively. The mean IC50 concentration of gefitinib in HCC827GR cells was decreased from 8,777 to 209.3 nM, and the mean IC50 concentration of osimertinib in PC9OR cells was decreased from 2,327 to 1,428 nM (Figure 7A-7C).
Figure 7.
RSL3 enhanced the cytotoxicity of gefitinib/osimertinib in LUAD EGFR-TKI-resistant cells. (A) Proliferation inhibition curves of HCC827GR cells after treatment with gefitinib or RSL3 plus gefitinib. (B) Proliferation inhibition curves of PC9OR cells after treatment with osimertinib or RSL3 plus osimertinib. (C) Mean IC50 concentration changes of gefitinib/osimertinib combined with RSL3. (D,E) PI staining analysis of HCC827GR and PC9OR cells after exposure to EGFR-TKIs or RSL3 plus EGFR-TKIs. +, cells were treated with RSL3/gefitinib/osimertinib; −, cells were treated with DMSO of the same volume as RSL3/gefitinib/osimertinib. (F) Immunofluorescence revealed the aggregation of ROS in HCC827GR and PC9OR cells (magnification, ×100). Data are exhibited as the mean ± standard error based on at least three independent experiments. Significant differences were defined by one-way ANOVA with a Bonferroni post-hoc test. ***, P<0.001. LUAD, lung adenocarcinoma; EGFR, epidermal growth factor receptor; TKI, tyrosine kinase inhibitor; ANOVA, analysis of variance.
We also treated HCC827GR and PC9OR cells with low doses of RSL3 (0.5 µM), gefitinib (1.0 µM), and osimertinib (1.0 µM) for 16 hours. We observed that individual treatment of RSL3, gefitinib, or osimertinib did not significantly affect the cell viability of HCC827GR and PC9OR. However, the cell survival rates of HCC827GR and PC9OR were significantly decreased after treating with a combination of RSL3 together with gefitinib or osimertinib (Figure 7D,7E), while ROS accumulation was increased at the same time (Figure 7F). These results suggested that RSL3 combined with EGFR-TKI could inhibit the proliferation of EGFR-TKI-resistant LUAD cells.
Discussion
At present, there is a pressing need to investigate the mechanism of TKI resistance, in order to identify new therapeutic targets in LUAD treatment (10,16). Aberrations or dysregulation of ferroptosis, as a kind of RCD, has been reported to be involved in the tumorigenesis of multiple cancers, and influences patient survival (36-38). However, the role of ferroptosis in TKI resistance remains unclear.
GPX4, a ferroptosis-related factor, is the main enzyme that reduces lipid peroxides and inhibits cell ferroptosis. It plays a similar role to that of B-Cell CLL/Lymphoma 2 (Bcl-2) by inhibiting cell apoptosis. By targeting GPX4, we could easily induce cell death in ferroptosis-sensitive cells (35,39). Upon the discovery of up-regulation of GPX4 in LUAD, especially in TKI-resistant LUAD cell lines, we proceeded to investigate the potential of GPX4 as a therapeutic target in LUAD after TKI resistance.
According to IHC staining of LUAD tissue microarray in this study, GPX4 was found to be mainly expressed in the cytoplasm in tumor cells as well as alveolar and bronchial epithelium. The positive rate of GPX4 was 67.07% (55/82) in LUAD tumors and 71.95% (59/82) in normal lung tissues, while the overall expression of GPX4 in LUAD tumors was stronger than that in normal lung tissues (Figure 2E).
Lung tissue is consistently in an oxygen-rich environment and has a strong ability to withstand oxidative damage. Under these conditions, patients with high GPX4 expression in tumor cells can maintain the balance between oxidation and antioxidation to avoid cell death by ferroptosis. This may explain why the expression level of GPX4 in tumor tissues is closely related to the survival of LUAD patients. Based on our study, the higher the expression level of GPX4, the better the prognosis. Lower expression levels of GPX4 were associated with poorer survival. However, the role of GPX4 as a protective factor, resulting in a better prognosis for LUAD patients, still requires further elucidation or validation.
We used RSL3, a small molecule inhibitor of GPX4, and Erastin, a small molecule inhibitor targeting the glutamate/cystine antiporter (System Xc-), to target LUAD cell lines (PC9, HCC78, H3122, and HCC827), and found that both RSL3 and Erastin exerted dose-dependent killing effects on the above LUAD cells. The average IC50 of RSL3 was significantly lower than that of Erastin, which suggests that targeting GPX4 to induce cell death is more effective in LUAD. By using the PC9OR, HCC827GR, HCC78-CR, and H3122LR cell lines, we evaluated the therapeutic effect of targeting GPX4 in LUAD after TKI resistance. We observed that RSL3 and Erastin had a dose-dependent inhibitory effect on the above-mentioned TKI resistant-cells. Moreover, the inhibiting effect of RSL3 on the above drug-resistant cells was better than that of Erastin.
Although RSL3 inhibits GPX4, the expression of NRF2 could resist lipid oxidative damage and protect cells from death. We subsequently combined RSL3 with the corresponding TKI, and found that RSL3 could reverse TKI resistance in the osimertinib-resistant cell strain of PC9 (PC9OR), the gefitinib-resistant cell strain of HCC827 (HCC827GR), and the lorlatinib-resistant cell strain of H3122 (H3122LR) cells, but not in the crizotinib-resistant cell strain of HCC78 (HCC78CR) (Figure S2), and the difference was statistically significant. These findings showed that GPX4 was only up-regulated in PC9OR and HCC827GR (Figure 5B), and no expression changes of GPX4 were detected in H3122LR and HCC78CR. The occurrence of enhanced cytotoxicity in H3122LR cells after combining RSL3 with Lorlatinib was unexpected, and we might explore this mechanism in future research. Ma et al. and Ni et al. also demonstrated that inhibition of GPX4 can reverse the resistance of TKIs in non-small cell lung cancer, indicating that targeting GPX4 is indeed a promising therapeutic strategy for overcoming TKI resistance in tumors (40,41).
In this paper, we described the expression characteristics of ferroptosis-related factors in LUAD by using multi-omics data from the TCGA database, and isolated the key ferroptosis-related factor, GPX4, which was up-regulated in tumor tissues and significantly correlated with the prognosis of LUAD patients. The up-regulation of GPX4 was also detected in a variety of LUAD cell lines after EGFR-TKI resistance. By targeting GPX4 with the small compound, RSL3, EGFR-TKI resistance can be reversed by inducing cell ferroptosis. Taken together, these findings offer a new therapeutic target for EGFR-TKI resistance in LUAD.
Conclusions
In summary, we described the expression characteristics of ferroptosis-related factors in LUAD as well as their relationship with the prognosis of LUAD patients, and demonstrated that GPX4 can serve as a new therapeutic target for EGFR-TKI resistance in LUAD.
Supplementary
The article’s supplementary files as
Acknowledgments
The authors appreciate the academic support from the AME Lung Cancer Collaborative Group.
Funding: This work was supported by the 1.3.5 Project for Disciplines of Excellence, West China Hospital, Sichuan University (No. ZYGD18021 to Lunxu Liu, No. ZYJC18009 to Jiandong Mei).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Institutional Review Board of West China Hospital, Sichuan University (No. 2016-98) and informed consent was taken from all the patients.
Footnotes
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-318/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-318/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-318/coif). TS receives grants from AstraZeneca, Chugai Pharmaceutical, Boehringer Ingelheim, Novartis and MSD, and payments or honoraria for lectures, presentations, speakers bureaus, manuscript writing or educational events from AstraZeneca, Chugai Pharmaceutical, Boehringer Ingelheim, Novartis, MSD, Taiho Pharma, Daiichi-Sankyo, Ono Pharmaceutical, Bristol-Myers Squibb, Nippon Kayaku and Pfizer. The other authors have no conflicts of interest to declare.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- 2.Zheng M. Classification and Pathology of Lung Cancer. Surg Oncol Clin N Am 2016;25:447-68. 10.1016/j.soc.2016.02.003 [DOI] [PubMed] [Google Scholar]
- 3.Blandin Knight S, Crosbie PA, Balata H, et al. Progress and prospects of early detection in lung cancer. Open Biol 2017;7:170070. 10.1098/rsob.170070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel MI, Cheng I, Gomez SL. US lung cancer trends by histologic type. Cancer 2015;121:1150-2. 10.1002/cncr.29180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jonna S, Subramaniam DS. Molecular diagnostics and targeted therapies in non-small cell lung cancer (NSCLC): an update. Discov Med 2019;27:167-70. [PubMed] [Google Scholar]
- 6.Denisenko TV, Budkevich IN, Zhivotovsky B. Cell death-based treatment of lung adenocarcinoma. Cell Death Dis 2018;9:117. 10.1038/s41419-017-0063-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet 2017;389:299-311. 10.1016/S0140-6736(16)30958-8 [DOI] [PubMed] [Google Scholar]
- 8.Miller KD, Nogueira L, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin 2019;69:363-85. 10.3322/caac.21565 [DOI] [PubMed] [Google Scholar]
- 9.McPhail S, Johnson S, Greenberg D, et al. Stage at diagnosis and early mortality from cancer in England. Br J Cancer 2015;112 Suppl 1:S108-15. 10.1038/bjc.2015.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jotte RM, Spigel DR. Advances in molecular-based personalized non-small-cell lung cancer therapy: targeting epidermal growth factor receptor and mechanisms of resistance. Cancer Med 2015;4:1621-32. 10.1002/cam4.506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sordella R, Bell DW, Haber DA, et al. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science 2004;305:1163-7. 10.1126/science.1101637 [DOI] [PubMed] [Google Scholar]
- 12.Nirmala JG, Lopus M. Cell death mechanisms in eukaryotes. Cell Biol Toxicol 2020;36:145-64. 10.1007/s10565-019-09496-2 [DOI] [PubMed] [Google Scholar]
- 13.Galluzzi L, Vitale I, Aaronson SA, et al. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ 2018;25:486-541. 10.1038/s41418-017-0012-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galluzzi L, Bravo-San Pedro JM, Vitale I, et al. Essential versus accessory aspects of cell death: recommendations of the NCCD 2015. Cell Death Differ 2015;22:58-73. 10.1038/cdd.2014.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimada K, Skouta R, Kaplan A, et al. Global survey of cell death mechanisms reveals metabolic regulation of ferroptosis. Nat Chem Biol 2016;12:497-503. 10.1038/nchembio.2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fulda S. Tumor resistance to apoptosis. Int J Cancer 2009;124:511-5. 10.1002/ijc.24064 [DOI] [PubMed] [Google Scholar]
- 17.Carneiro BA, El-Deiry WS. Targeting apoptosis in cancer therapy. Nat Rev Clin Oncol 2020;17:395-417. 10.1038/s41571-020-0341-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dixon SJ, Lemberg KM, Lamprecht MR, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 2012;149:1060-72. 10.1016/j.cell.2012.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie Y, Hou W, Song X, et al. Ferroptosis: process and function. Cell Death Differ 2016;23:369-79. 10.1038/cdd.2015.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang WS, Stockwell BR. Ferroptosis: Death by Lipid Peroxidation. Trends Cell Biol 2016;26:165-76. 10.1016/j.tcb.2015.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mou Y, Wang J, Wu J, et al. Ferroptosis, a new form of cell death: opportunities and challenges in cancer. J Hematol Oncol 2019;12:34. 10.1186/s13045-019-0720-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao X, Tang M, Tian S, et al. A ferroptosis-related gene signature predicts overall survival in patients with lung adenocarcinoma. Future Oncol 2021;17:1533-44. 10.2217/fon-2020-1113 [DOI] [PubMed] [Google Scholar]
- 23.Liang JY, Wang DS, Lin HC, et al. A Novel Ferroptosis-related Gene Signature for Overall Survival Prediction in Patients with Hepatocellular Carcinoma. Int J Biol Sci 2020;16:2430-41. 10.7150/ijbs.45050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vasan N, Baselga J, Hyman DM. A view on drug resistance in cancer. Nature 2019;575:299-309. 10.1038/s41586-019-1730-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim ZF, Ma PC. Emerging insights of tumor heterogeneity and drug resistance mechanisms in lung cancer targeted therapy. J Hematol Oncol 2019;12:134. 10.1186/s13045-019-0818-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hassannia B, Vandenabeele P, Vanden Berghe T. Targeting Ferroptosis to Iron Out Cancer. Cancer Cell 2019;35:830-49. 10.1016/j.ccell.2019.04.002 [DOI] [PubMed] [Google Scholar]
- 27.Lei G, Zhang Y, Koppula P, et al. The role of ferroptosis in ionizing radiation-induced cell death and tumor suppression. Cell Res 2020;30:146-62. 10.1038/s41422-019-0263-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prasad S, Gupta SC, Tyagi AK. Reactive oxygen species (ROS) and cancer: Role of antioxidative nutraceuticals. Cancer Lett 2017;387:95-105. 10.1016/j.canlet.2016.03.042 [DOI] [PubMed] [Google Scholar]
- 29.Srinivas US, Tan BWQ, Vellayappan BA, et al. ROS and the DNA damage response in cancer. Redox Biol 2019;25:101084. 10.1016/j.redox.2018.101084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hayes JD, Dinkova-Kostova AT, Tew KD. Oxidative Stress in Cancer. Cancer Cell 2020;38:167-97. 10.1016/j.ccell.2020.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stockwell BR, Friedmann Angeli JP, Bayir H, et al. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell 2017;171:273-85. 10.1016/j.cell.2017.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang C, Zhang X, Yang M, et al. Recent Progress in Ferroptosis Inducers for Cancer Therapy. Adv Mater 2019;31:e1904197. 10.1002/adma.201904197 [DOI] [PubMed] [Google Scholar]
- 33.Detre S, Saclani Jotti G, Dowsett M. A. "quickscore" method for immunohistochemical semiquantitation: validation for oestrogen receptor in breast carcinomas. J Clin Pathol 1995;48:876-8. 10.1136/jcp.48.9.876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Yan H, Xu X, et al. Erastin/sorafenib induces cisplatin-resistant non-small cell lung cancer cell ferroptosis through inhibition of the Nrf2/xCT pathway. Oncol Lett 2020;19:323-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang WS, SriRamaratnam R, Welsch ME, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell 2014;156:317-31. 10.1016/j.cell.2013.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Friedmann Angeli JP, Krysko DV, Conrad M. Ferroptosis at the crossroads of cancer-acquired drug resistance and immune evasion. Nat Rev Cancer 2019;19:405-14. 10.1038/s41568-019-0149-1 [DOI] [PubMed] [Google Scholar]
- 37.Sun X, Ou Z, Chen R, et al. Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology 2016;63:173-84. 10.1002/hep.28251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fonseca-Nunes A, Jakszyn P, Agudo A. Iron and cancer risk--a systematic review and meta-analysis of the epidemiological evidence. Cancer Epidemiol Biomarkers Prev 2014;23:12-31. 10.1158/1055-9965.EPI-13-0733 [DOI] [PubMed] [Google Scholar]
- 39.Bersuker K, Hendricks JM, Li Z, et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature 2019;575:688-92. 10.1038/s41586-019-1705-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma CS, Lv QM, Zhang KR, et al. NRF2-GPX4/SOD2 axis imparts resistance to EGFR-tyrosine kinase inhibitors in non-small-cell lung cancer cells. Acta Pharmacol Sin 2021;42:613-23. 10.1038/s41401-020-0443-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ni J, Chen K, Zhang J, et al. Inhibition of GPX4 or mTOR overcomes resistance to Lapatinib via promoting ferroptosis in NSCLC cells. Biochem Biophys Res Commun 2021;567:154-60. 10.1016/j.bbrc.2021.06.051 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as