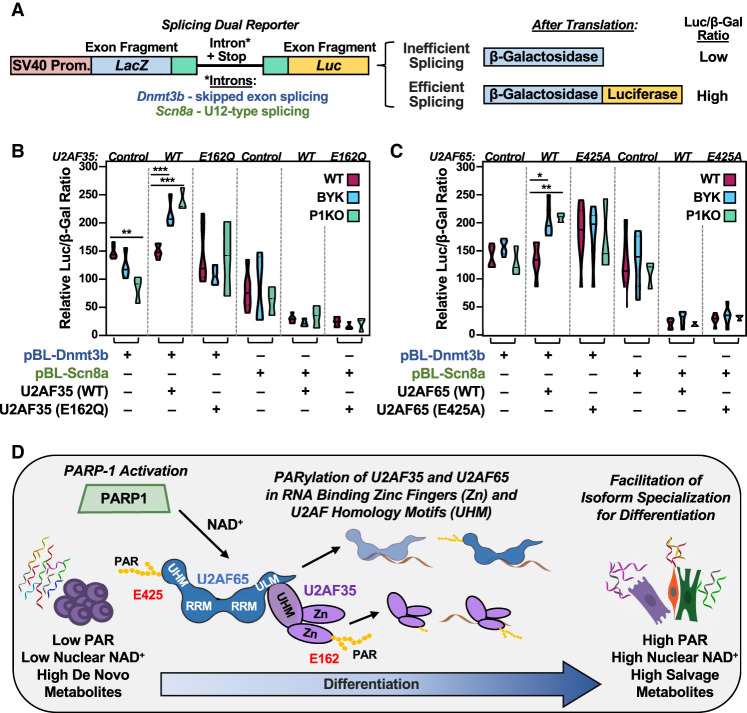
Figure 7.
Inhibition of PARP1 catalytic activity affects U2AF splicing functions during the differentiation of mESCs. (A) Diagram of a pBPLUGA in vivo dual-splicing reporter containing fragments of splice sites of the Dnmt3b (U2-type splicing) and Scn8a (U12-type splicing) genes. The reporter contains two reporter genes (LacZ encoding β-galactosidase and Luc encoding luciferase). Efficient splicing removes in-frame stop codons in the intron, allowing for expression of a LacZ + Luc fusion. (B,C) Relative luciferase/β-galactosidase activity ratios from assays in WT and P1KO mESCs using the Dnmt3b and Scn8a pBPLUGA constructs in the presence of WT or ADPRylation site mutant U2AF (E162Q U2AF35 and E425A U2AF65). The reporter and U2AF35 or U2AF65 expression plasmids were electroporated into mESCs. The mESCs were then differentiated for 12 h by LIF removal. In some cases, as indicated, the cells were treated with the PARP inhibitor BYK204165 at 10 µM for 2 h prior to collection. Summary of results from multiple experiments displayed as violin plots (n ≥ 3, statistical tests were performed using two-way ANOVA and multiple comparisons within each group of three conditions [WT, BYK, and P1KO]; Sidak). (∗) P < 0.05, (∗∗) P < 0.01, (∗∗∗) P < 0.001. (D) Model showing the regulation of embryonic stem cell state by the NAD+–PARP1 axis through site-specific PARylation of the U2AF complex proteins U2AF35 and U2AF65, resulting in altered splicing of mRNAs encoding key embryonic stem cell state proteins.