Fig. 4. Short arms of the acrocentric chromosomes.
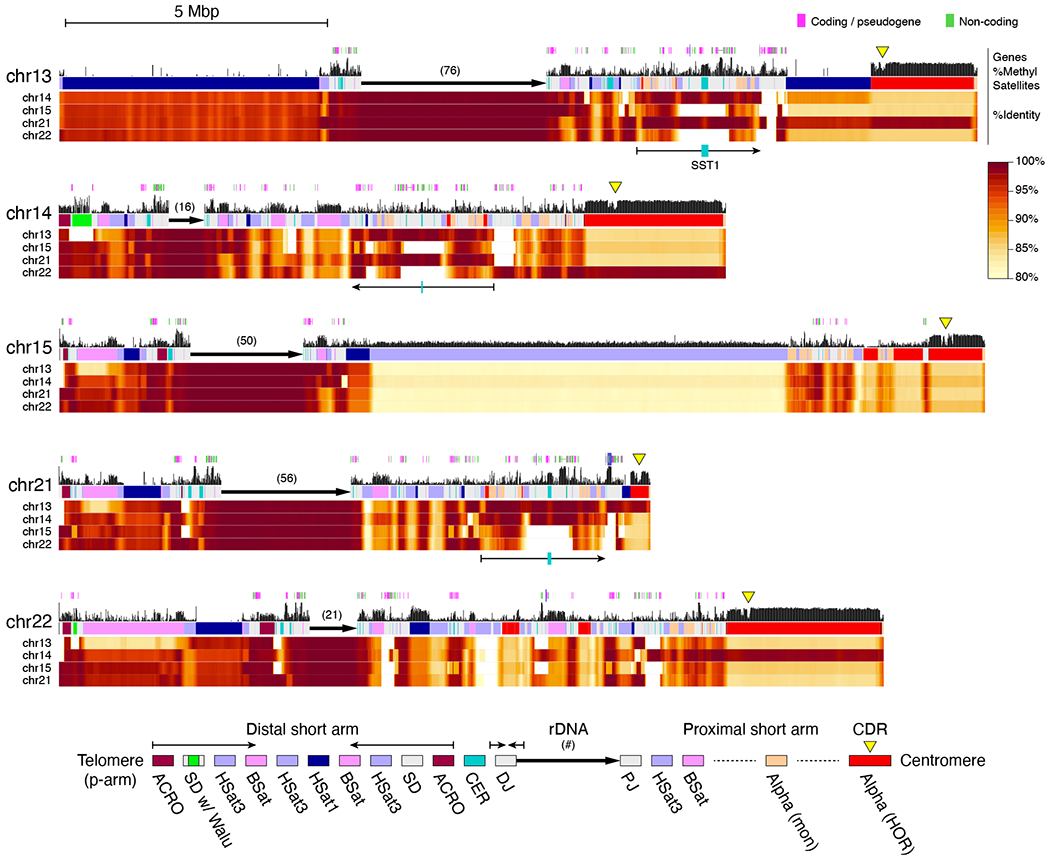
Each short arm is shown along with annotated genes, percent of methylated CpGs (29), and a color-coded satellite repeat annotation (30). The rDNA arrays are represented by a directional arrow and copy number due to their high self-similarity, which prohibits ONT mapping. Percent identity heatmaps versus the other four arms were computed in 10 kbp windows and smoothed over 100 kbp intervals. Each position shows the maximum identity of that window to any window in the other chromosome. The distal short arms include conserved satellite structure and inverted repeats (thin arrows), while the proximal short arms show a diversity of structures. The proximal short arms of Chromosomes 13, 14, and 21 share a segmentally duplicated core, including small alpha satellite HOR arrays and a central, highly methylated, SST1 array (thin arrows with teal block). Yellow triangles indicate hypomethylated centromeric dip regions (CDRs), marking the sites of kinetochore assembly (29).
