Fig. 5. Resolved FRG1 paralogs.
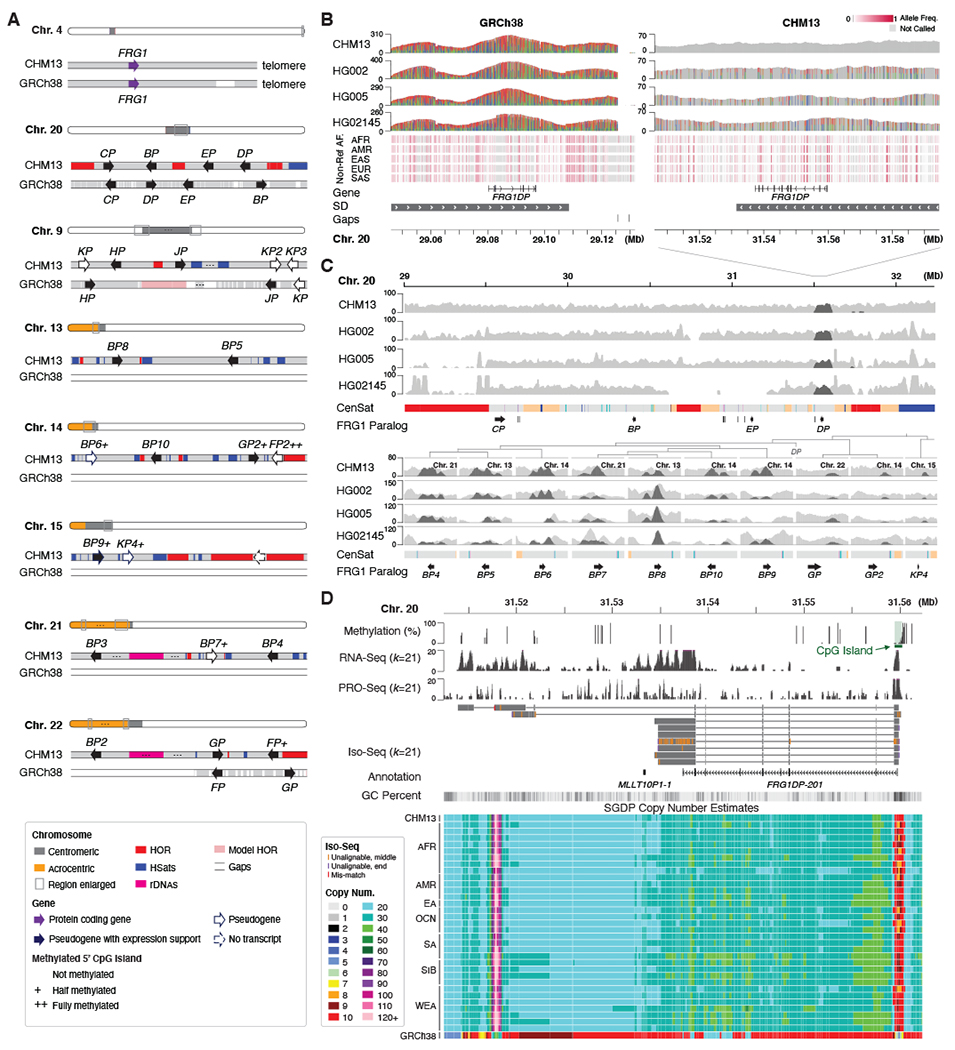
(A) Protein-coding gene FRG1 and its 23 paralogs in CHM13. Only 9 are found in GRCh38. Genes are drawn larger than their actual size and the “FRG1” prefix is omitted for brevity. All paralogs are found near satellite arrays. Most copies exhibit evidence of expression, including CpG islands present at the 5′ start site with varying degrees of methylation. (B) Reference (gray) and variant (colored) allele coverage is shown for four human HiFi samples mapped to the paralog FRG1DP. When mapped to GRCh38, the region shows excessive HiFi coverage and variants, indicating that reads from the missing paralogs are mis-mapped to FRG1DP (variants with >80% coverage shown). When mapped to CHM13, HiFi reads show the expected coverage and a typical heterozygous variation pattern for the three non-CHM13 samples (variants >20% coverage shown). These non-reference alleles are also found in other populations from 1KGP ILMN data. (C) Mapped HiFi read coverage for other FRG1 paralogs, with an extended context shown for Chromosome 20. Coverage of HiFi reads that mapped to FRG1DP in GRCh38 are highlighted (dark gray), showing the paralogous copies they originate from (FRG1BP4–10, FRG1GP, FRG1GP2, and FRG1KP4). Background coverage is variable for some paralogs, suggesting copy number polymorphism in the population. (D) Methylation and expression profiles suggest transcription of FRG1DP in CHM13. In the copy number display (bottom), each length k sequence (k-mer) of the CHM13 assembly is painted with a color representing the copy number of that k-mer sequence in an SGDP sample. The CHM13 and GRCh38 tracks show the copy number of these same k-mers in the respective assemblies. CHM13 copy number resembles all samples from the SGDP, whereas GRCh38 underrepresents the true copy number.
