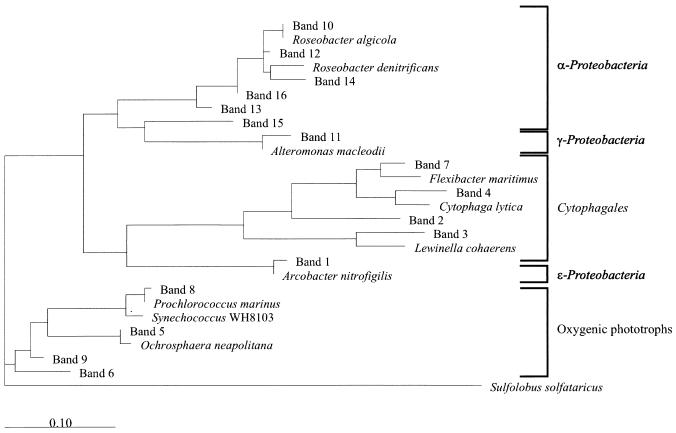
FIG. 5.
Phylogenetic tree showing relationships of the sequences found in our samples to representative bacterial 16S rRNA genes. Sequences of the cultured bacteria were retrieved from GenBank. The tree was inferred by the maximum-likelihood method using full-length genes for the cultured bacteria and the entire length of the sequenced bands (169 to 194 bp) beginning at the equivalent to base 341 and going towards the 3′ end of the 16S rRNA molecule (Escherichia coli numbers). An Archaea (Sulfolobus solfataricus) was used as an outgroup. GenBank accession numbers of the cultured bacteria are as follows: R. algicola, X78314; R. denitrificans, M96746; A. macleodii, Y18228; F. maritimus, M64629; C. lytica, M62796; L. cohaerens, AF039292; A. nitrofigilis, L14627; P. marinus, X63140; Synechococcus sp. strain WH8103, AF001479; O. neapolitana, X80390. The scale bar indicates substitutions per nucleotide position.