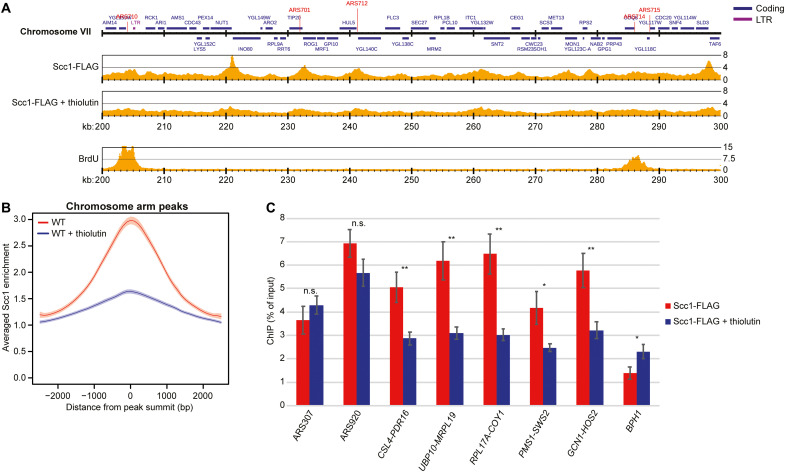
Fig. 3. Transcription inhibition delocalizes dynamic cohesin from its normal binding sites between convergently oriented gene pairs.
(A) Scc1-FLAG enrichment on chromosome VII (200 to 300 kb from left telomere) based on normalized ChIP-seq analysis of untreated and thiolutin-treated WT cells arrested in S phase by HU (two upper maps). The lowest map shows 5-bromo-2′-deoxyuridine (BrdU) incorporation in the same chromosomal region in synchronized WT cells arrested in S phase by HU (23). Note that replication forks have progressed a few kilobase pairs from the early firing origins ARS710 and ARS714, leaving most of the region in view unreplicated. The y axis shows fold enrichment of ChIP / input in linear scale, and the x axis shows chromosomal positions. Blue and purple horizontal bars in the uppermost genomic region panel denote coding regions and long terminal repeats (LTRs), respectively, and red vertical lines indicate replication origins [autonomously replicating sequence (ARS)]. (B) Averaged Scc1-FLAG enrichment at cohesin sites along chromosome arms, based on the analysis presented in (A). Shaded regions indicate the 95% confidence interval. (C) Chromosomal association of Scc1-FLAG at two early replication origins (ARS307 and ARS920) at selected cohesin sites (intergenic regions indicated by flanking convergently oriented gene pairs), and within the 5′ end of BPH1 ORF, as determined by ChIP-qPCR of samples collected from cells treated as in (A). N = 3, n.s. P > 0.05, *P ≤ 0.05, **P ≤ 0.01.