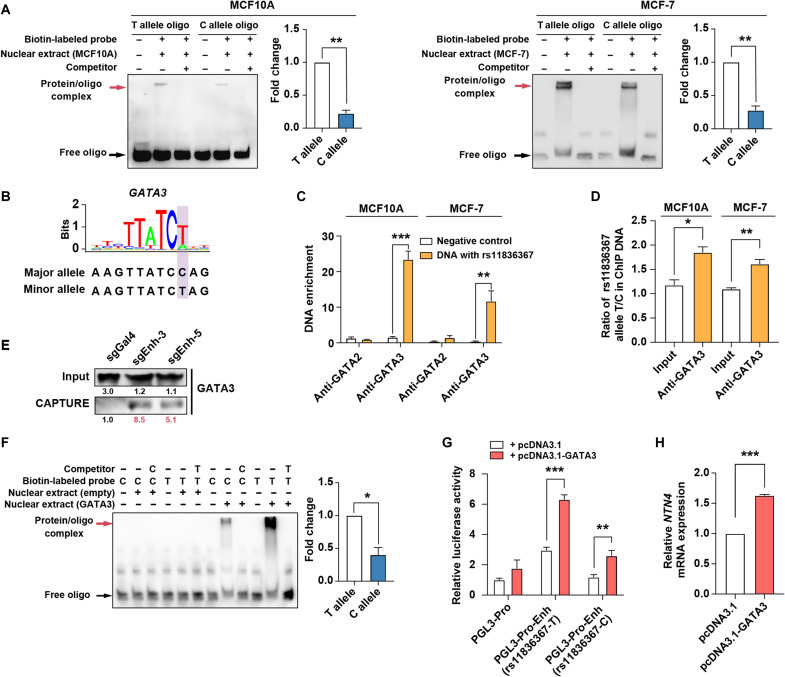
Fig. 2. rs11836367 alleles specifically influence GATA3 binding affinity.
(A) The binding affinities between oligos containing the C allele or T allele of rs11836367 and nuclear extracts (from MCF10A and MCF-7 cells) were analyzed by EMSA, and shifted bands were quantified. (B) The risk C allele of rs11836367 interferes with a GATA3 binding motif. (C) ChIP-qPCR for GATA2 and GATA3 at the DNA fragment containing rs11836367 locus. (D) Allele-specific ChIP-qPCR for GATA3 at the DNA fragment containing rs11836367 locus. (E) Western blot showing enrichment of GATA3 at the rs11836367-containing enhancer in MCF10A by in situ CAPTURE assay. sgEnh-3 and sgEnh-5 target the enhancer (containing rs11836367), and sgGal4 is a negative control. (F) The binding affinities between oligos containing the C allele or T allele of rs11836367 and GATA3 were analyzed by EMSA (left), and shifted bands were quantified (right). Nuclear extracts were obtained from 293T cells transfected with either an empty vector or GATA3-overexpressing construct. (G) Luciferase reporter assay performed after transient cotransfection of MCF10A cells with the indicated constructs. The DNA fragment containing either the T or C allele of rs11836367 was cloned into the NTN4 promoter (PGL3-Pro)–driven luciferase reporter construct (PGL3-Pro-Enh-rs11836367-T vector or PGL3-Pro-Enh-rs11836367-C vector), which was cotransfected with either an empty vector (pcDNA3.1) or a GATA3-overexpressing vector (pcDNA3.1-GATA3). (H) qRT-PCR showing the transfection of MCF10A cells with pcDNA3.1-GATA3 increased NTN4 mRNA expression. Data are represented as means ± SEM of three independent experiments. *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001.