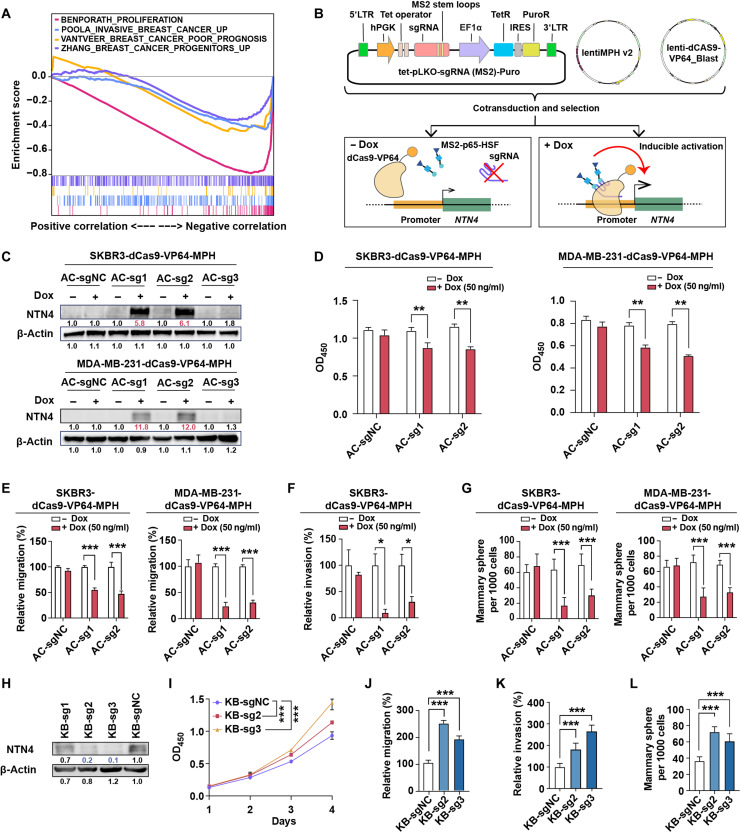
Fig. 4. NTN4 inhibits proliferation, migration, invasion, and cancer stem cell features in breast cancer cells.
(A) Gene set enrichment analysis (GSEA) analysis of genes preranked by Pearson correlation to NTN4 expression for four breast cancer–related signatures, which were BENPORATH_PROLIFERATIION [red curve: normalized enrichment score (NES) = −3.87, false discovery rate (FDR) q value < 0.001], POOLA_INVASIVE_BREAST_CANCER_UP (blue curve: NES = −2.34, FDR q value < 0.001), VANTVEER_BREAST_CANCER_POOR_PROGNOSIS (yellow curve: NES = −1.81, FDR q value = 0.001), and ZHANG_BREAST_CANCER_PROGENITORS_UP (purple curve: NES = −2.01, FDR q value = 0.001) in RNA sequencing (RNA-seq) datasets from TCGA. (B) Schematic of dCas9-SAM–mediated doxycycline (Dox)–inducible endogenous overexpression of NTN4. TetR, Tet repressor; IRES, internal ribosomal entry site; 5’LTR, 5’ long terminal repeat; PuroR, puromycin resistent gene. (C) Validation of inducible overexpression of the NTN4 protein by Western blot analysis in SKBR3-dCas9-VP64-MPH and MDA-MB-231-dCas9-VP64-MPH cells following the addition of Dox (50 ng/ml). AC-sg1, AC-sg2, and AC-sg3 were designed to target the NTN4 promoter, and notargeting AC-sgNC was a negative control. (D) Cell proliferation was analyzed by Cell Counting Kit-8 (CCK-8) after 4-day culture with or without Dox treatment. OD450, optical density at 450 nm. (E) Cellular migration was analyzed using the Transwell assay with or without Dox treatment. (F) Cellular invasion was investigated using the three-dimensional spheroid invasion assay in Matrigel following a 6-day culture with or without Dox treatment. (G) Cancer stem cell features were analyzed using sphere formation assays after a 7-day culture with or without Dox treatment. (H) Validation of CRISPRi knockdown of NTN4 protein by Western blot analysis in SUM159-dCas9-KRAB cells. KB-sg1, KB-sg2 and KB-sg3 were designed to target the NTN4 promoter, and nontargeting KB-sgNC was used as a negative control. (I to L) Cellular proliferation (I), migration (J), invasion (K), and cancer stem cell features (L) were analyzed in NTN4 knockdown SUM159-dCas9-KRAB cell lines as described above. Data are represented as means ± SEM of three to six independent experiments. *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001. Related representative images for (E) to (G) and (J) to (L) were shown in fig. S4.