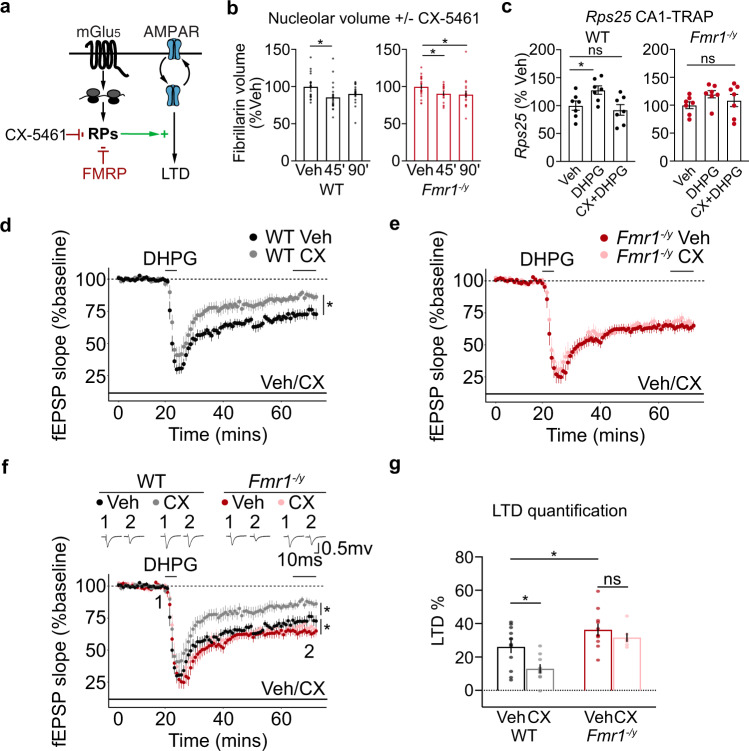
Fig. 3. RP translation is required for mGluR-LTD in WT but not in Fmr1−/y hippocampal CA1.
a Schematic of experimental strategy. Specific RNA Pol1 blocker CX-5461 was used to ribogenesis, including RP translation. b Fibrillarin staining of hippocampal slices shows that nucleolar volume of CA1 pyr neurons is significantly reduced in both genotypes with 200 nM CX-5461 treatment by 45 min (WT: Veh = 100 ± 3.0%, CX 45’ = 85.9 ± 3.8%, CX 90’ = 90.8 ± 2.7%. One-way ANOVA *P = 0.0206, post hoc Veh-45’ *FDR = 0.0141, Veh-90’ FDR = 0.070. N = 20 neurons per group). (Fmr1−/y: Veh = 100 ± 2.5%, CX 45’ = 90.96 ± 2.4%, CX 90’ = 90.05 ± 4.2%. One-way ANOVA *P = 0.0488, post hoc *Veh-45’ FDR = 0.0485, *Veh-90’ FDR = 0.0485, N = 20 neurons per group). c CA1-TRAP and qPCR analysis of Rps25 translation in hippocampal slices confirms an upregulation in the WT in response to DHPG at 30’ that is blocked with 200 nM CX-5461. DHPG does not elicit a significant increase in Rps25 translation in Fmr1−/y slices nor does CX-5461 have a significant effect (WT: Veh = 100 ± 8.22%, DHPG = 127.9 ± 7.5%, CX + DHPG = 92.3 ± 9.5%. One-way ANOVA *P = 0.0388, post hoc Veh-DHPG *FDR = 0.0121, Veh-CXDHPG FDR = 0.3506, N = 7 littermate pairs. Fmr1−/y: Veh = 100 ± 6.2%, DHPG = 119.6 ± 6.5%, CX + DHPG = 108.5 ± 10.9%. One-way ANOVA P = 0.2627). N = 7 littermate pairs. Data are presented as mean values + /− SEM. d, e mGluR-LTD was measured in hippocampal CA1 in the presence of vehicle or CX-5461. Slices were prepared from Fmr1−/y and WT, recovered for at least 2 h, and exposed to vehicle or 200 nM CX-5461 for at least 30 min prior to DHPG application and throughout the LTD recording. CX-5461 causes a significant reduction in LTD magnitude in WT CA1 versus vehicle (Veh = 26.13% ± 3.5% N = 12 animals, CX-5461 = 13.00% ± 2.4% N = 10 animals, *FDR = 0.0031). In contrast, CX-5461 does not significantly reduce LTD magnitude in Fmr1−/y CA1 versus vehicle (Veh = 36.337% ± 3.9% N = 11 animals, CX-5461 = 31.717% ± 2.3% N = 7, animals. FDR = 0.1224). f, g Quantification of all four groups show a differential effect of CX-5461 on LTD in each genotype (WT-Veh = 26.13% ± 3.5% N = 12 animals, WT-CX54611 = 13.00% ± 2.4% N = 10 animals, Fmr1−/y Veh = 36.337% ± 3.9% N = 11 animals, Fmr1−/y CX-5461 = 31.717% ± 2.3% N = 7, animals. Two-way ANOVA, genotype *P < 0.001 treatment *P = 0.01, post hoc: WT-Veh vs Fmr1−/y-Veh *FDR = 0.0107, WT-Veh vs WT-CX-5461 *FDR = 0.0031, Fmr1−/y-Veh vs Fmr1−/y-CX-5461 FDR = 0.1224). Data are presented as mean values + /− SEM. Source data are provided as a Source Data file.