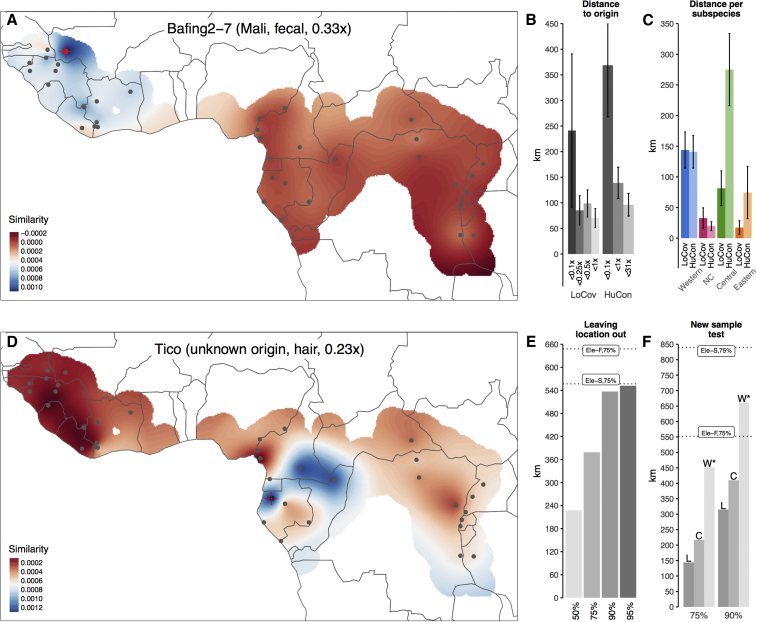
Figure 4.
Chimpanzee geolocalization based on rare variation
(A) Spatial model of shared rare alleles with 38 sampling locations. Red indicates lower amounts, while blue indicates larger amounts of shared rare alleles. Black dots, locations included in the reference panel; red dot, known place of origin (low-coverage sample Baf2-7 from Bafing, Mali); red cross, inferred origin. Red cross and dot overlap in this correctly assigned sample.
(B) Average distance (km) of best matching to true location in bins of coverage for samples with low coverage and contamination (LoCov) (n = 99) and samples with human contamination of more than 0.5% (HuCon) (n = 139). Error bars represent the SEM.
(C) Average distance of best matching to true location per subspecies, stratified by low-coverage and human-contaminated samples; note that the Nigeria-Cameroon chimpanzee range is smaller than that of other subspecies, thus resulting in smaller distances. Error bars represent the SEM.
(D) Geolocalization of the chimpanzee Tico from a rescue center in Spain, here assigned to Gabon or Equatorial Guinea.
(E) Assignment accuracy when leaving full locations out (n = 434), with 50th, 75th, 90th, and 95th percentiles for the distance of inferred to true origin; for comparison, best 75th percentiles for geolocalization of elephants21 are shown as dotted lines (Ele-S, Savanna elephant; Ele-F, forest elephant).
(F) Assignment accuracy for samples not included in the reference panel (L, low coverage; C, contaminated; W, whole-genome data6); for comparison, the 75th percentiles of the single sample test in elephants are shown as dotted lines; the asterisk marks that the origin for whole genomes may be different from the place of confiscation reported for these individuals. See also Figures S67–S75, S80, and S81.