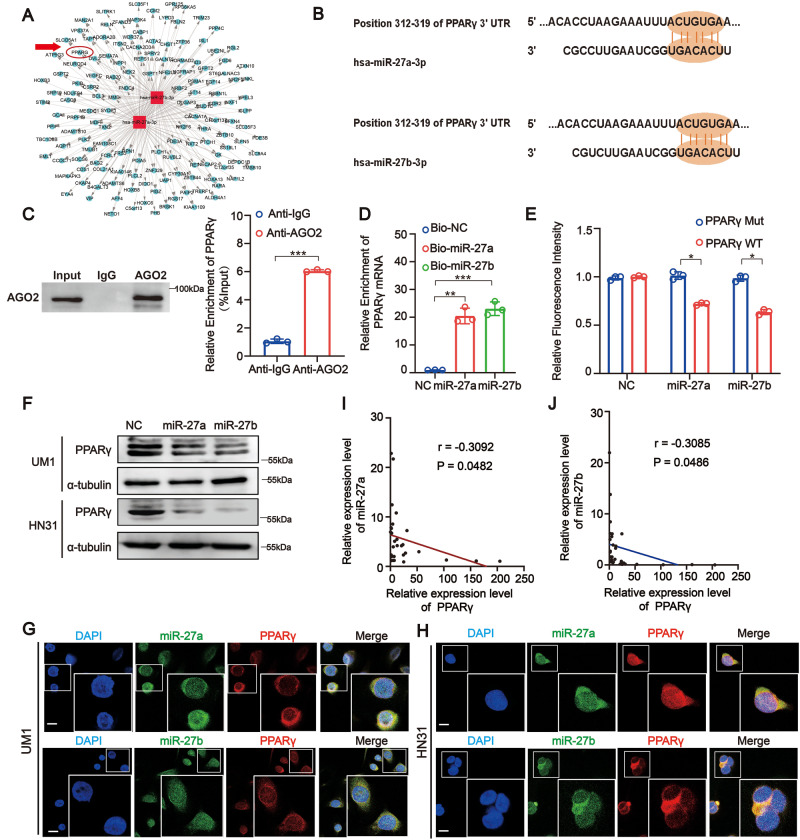
Fig. 5. PPARγ is a direct target of both miR-27a and miR-27b in HNSCC.
A The network of target genes of miR-27a and miR-27b, the red circle represents PPARγ. B Schematic illustration showed binding site sequences between PPARγ and miR-27a/27b. C RIP assay showed PPARγ mRNA was effectively pulled down by AGO2 with a much greater enrichment in UM1 cells. D RNA pull-down assay exhibited for the direct bonding of PPARγ and miR-27a/27b in UM1 cells. E Dual-luciferase assays were conducted using PPARγ wild type (WT) and mutant (MUT) 3′UTR reporter constructs to show that the human PPARγ 3′UTR region contains miR-27a and miR-27b binding sites. (Data were normalized to that with the control probe, showing the mean ± SD of three experiments). F Western Blot assay showed miR-27a and miR-27b mimic could significantly reduce expression of PPARγ in UM1 and HN31 cells at the level of proteins. G, H RNA FISH showed colocalization of PPARγ and miR-27a/27b in UM1 and HN31 cells (Scale bar, 10 μm). I, J Pearson correlation analysis indicated miR-27a and miR-27b expression was negatively correlated with PPARγ expression in patients with HNSCC (n = 15 and 15, respectively). Data were shown as the mean value ± SD of three independent experiments. The asterisks indicate significant differences (Student’s t-tests, *P < 0.05, **P < 0.01, ***P < 0.001).