Figure 1.
Identification of conserved and non-conserved antibody epitopes recognized following COVID-19 vaccination
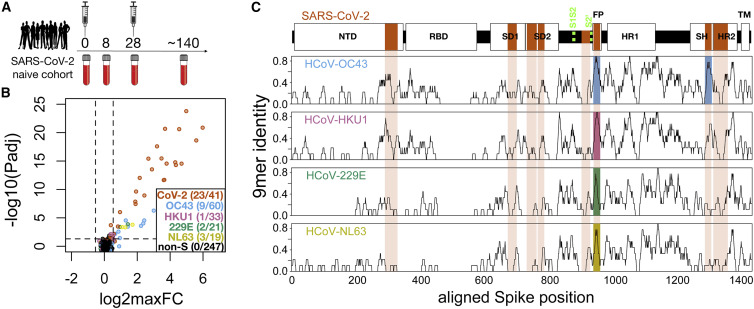
(A) A cohort of 21 subjects with no prior SARS-CoV-2 infection history and undetectable SARS-CoV-2 antibodies prior to vaccination received two doses of the mRNA-1273 vaccine and gave blood samples at 0, ∼8, ∼28, and ∼140 days relative to the first dose.
(B) Plasma from each time point was analyzed for IgG reactivity by PepSeq with a 15,000-peptide human virome library (“HV2”). Z scores for the 421 HV2 peptides designed from members of the Coronaviridae family were analyzed across subjects and time points to identify peptides showing significant time-differential signal. Each dot represents an individual peptide: the y axis shows the FDR-adjusted ANOVA p value (−log10Padj) across all time points, and the x axis shows the maximum log2 fold change (log2maxFC), which was calculated by dividing the maximum Z score at days 8, 28, and 140 by the Z score at day 0. The key in the lower right shows the number of peptides for each CoV species (note: HCoV-OC43 is a member of the Beta-CoV-1 species and SARS-CoV-2 is a member of the SARSr-CoV species). Dashed lines indicate thresholds at Padj = 0.05 and maxFC = 1.45.
(C) Mapping of differentially recognized peptides (passing both Padj and maxFC thresholds shown in [B]) to a multiple sequence alignment of Spike proteins from SARS-CoV-2, HCoV-OC43, HCoV-HKU1, HCoV-229E, and HCoV-NL63. Plots show structural features or cleavage sites (dotted green lines) of SARS-CoV-2 Spike (top) and sliding-9-mer window amino acid sequence identity to SARS-CoV-2 Spike for the endemic Spike proteins (bottom). The differentially recognized peptides map to eight epitopes in SARS-CoV-2 Spike (vertical orange boxes), two epitopes in HCoV-OC43 (blue), one epitope in HCoV-HKU1 (pink), one epitope in HCoV-229E (green), and one epitope in HCoV-NL63 (gold). Epitopes detected in the endemic CoV proteins all occur at two regions of high sequence conservation with SARS-CoV-2. The following features of Spike are highlighted: NTD, N-terminal domain; RBD, receptor binding domain; SD1, subdomain 1; SD2, subdomain 2; FP, fusion peptide; HR1, heptad repeat 1; SH, stem helix; HR2, heptad repeat 2; TM, transmembrane region.