Figure 5.
Responses to FP and SH epitopes depend on both conserved and polymorphic residues
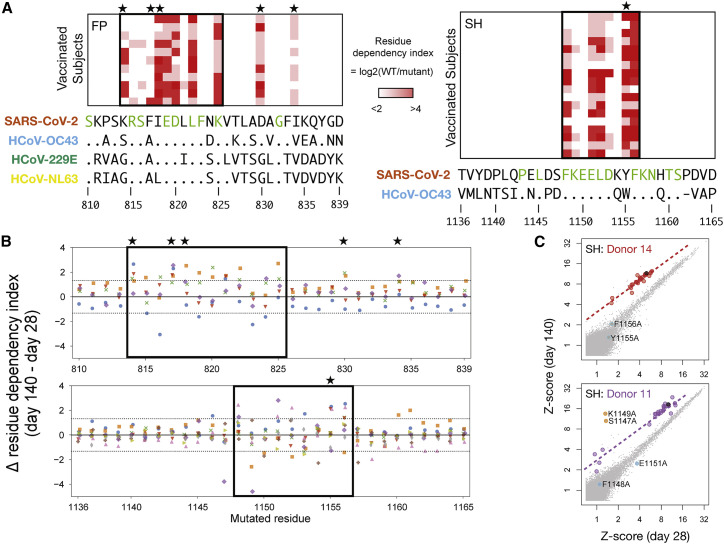
(A) PepSeq analysis of vaccinated subjects (rows) at day 140 for sets of HV2 peptides (columns) in which each residue of SARS-CoV-2 Spike at positions 810–839 (containing FP region, left) or positions 1,136–1,165 (containing SH region, right) was individually substituted to alanine. Color scale indicates the log2(ratio) of PepSeq Z scores in each subject for the wild-type peptide, divided by the respective alanine-substituted peptide (“residue dependency index”). Boxes enclose the inferred core binding regions, and the sequence letters colored in green below indicate positions that are conserved between all species shown. Stars indicate polymorphic positions that are involved in binding. Shown are subjects for whom the response to the respective wild-type peptide exceeds a Z score threshold of 10: respectively, 11 and 18 subjects for FP and SH.
(B) Analysis of the alanine-substituted FP (top) and SH (bottom) peptide sets described in (A), now comparing the responses at day 28 versus day 140 for subjects (each in a different color) whose reactivity to the wild-type peptides at both time points exceeded a Z score threshold of 10: respectively, 5 and 9 subjects for FP and SH. The y axis shows the log2 change (from the day 28 to the day 140 time point) in residue dependency (as defined in [A]), at the position shown on the x axis. Dotted horizontal lines demarcate the 99th percentile range of variation observed in a set of 1,830 control data points from the same nine subjects at 11 non-CoV epitopes to which stable (non-vaccine-induced) responses were detected and that were also each represented as sets of 30 alanine mutants and analyzed in the same way.
(C) Example scatterplots showing peptide-level reactivity at day 28 versus day 140 in two subjects with evidence for temporal changes in residue-level specificity within the SH epitope. Alanine-mutated SH peptides are highlighted in red/purple (corresponding to their respective donor’s color in [B]), the wild-type SH peptide in black, and non-SH peptides in gray. Peptides with significant temporal deviation relative to wild type (as calculated in [B]) are shown in cyan (increasing dependency on the substituted position) or orange (decreasing dependency on the substituted position) and annotated with the coordinate of the substituted residue. Dashed lines show the fit for mutant peptides without significant temporal deviation.