Fig. 4. Comparison of the structures of class A muscarinic M2R, class B Glucagon GCGR, and class C metabotropic glutamate mGlu2R in complex with Gi or arrestin-2.
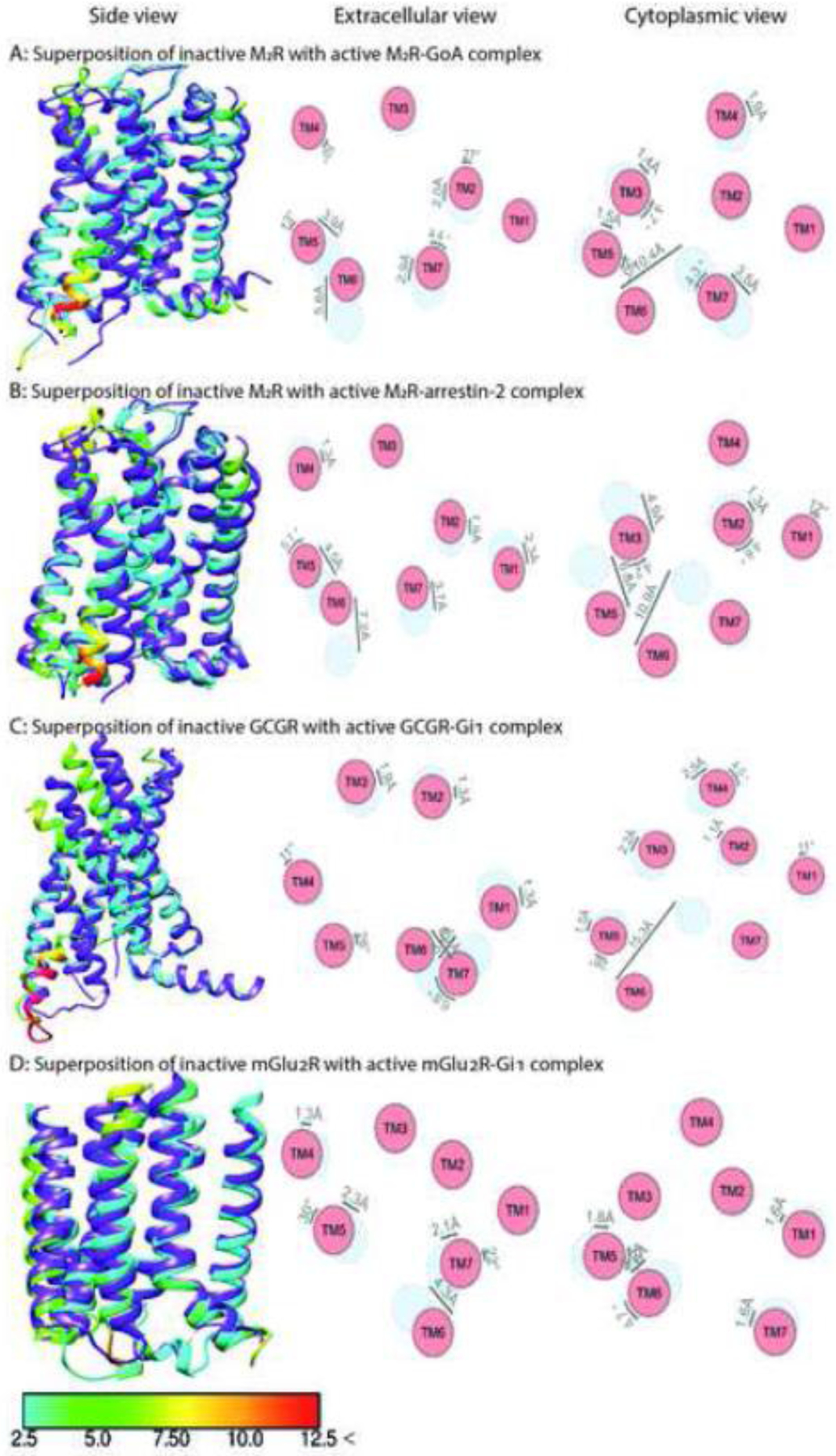
The structure of inactive M2R (PDB: 3UON) [88] is superposed with activated M2R in complex with GoA (panel A, PDB: 6OIK) [40] and arrestin-2 (panel B, PDB: 6U1N) [46]. The structure of inactive GCGR (PDB: 5YQZ) [87] is superposed with activated GCGR in complex with Gi1 (panel C, PDB: 6LML) [42]. The structure of inactive mGlu2R (PDB: 7MTQ) [44] is superposed with activated mGlu2R in complex with Gi1 (panel D, PDB: 7MTS) [44]. Shown are side views (left panels), and schematic representation of helices from extracellular (middle panels) and cytoplasmic views (right panel). The positions of helices in the inactive state are shown in light blue, active in red. Inactive receptors are colored purple in left panels. The residues of the active receptor in complex with intracellular partners are color coded based on the RMSD of α-carbons from the matched residues in the inactive receptor with thresholds of 2.5 Å (cyan), 5 Å (green), 7.5 Å (yellow), 10 Å (orange), and 12.5 Å (red). The direction and extent of displacement of helices are also indicated by arrows.
