Figure 2. Structural basis for site-specific ubiquitylation of nucleosomal H2A.
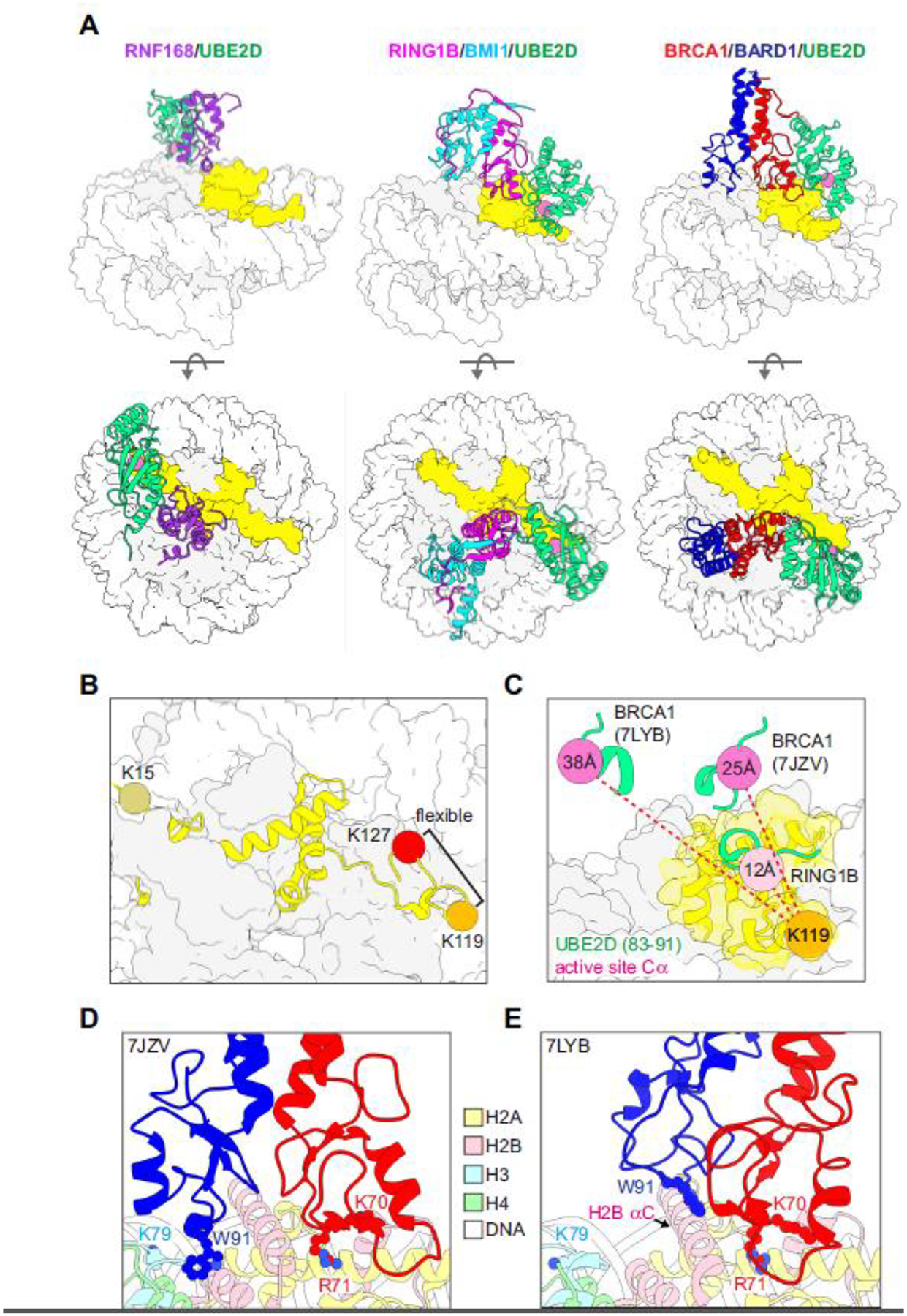
(A) Structural models of RING/E2/nucleosome complexes (PDB-Dev: 00000028, PDB: 4R8P, 7JZV). The active site Cα of the E2 (UBE2D, green) in each model is shown as a pink sphere and H2A is yellow. (B) Locations of representative target lysine residues for H2A-modifying Ub ligases on a nucleosome (PDB: 1KX5). The C-terminal tail of H2A beyond K119 is unstructured and not observed in x-ray or cryo-EM structures but is modelled in PDB 1KX5. (C) Positions of the E2 active site (Cα, pink spheres) from the indicated RING/E2/nucleosome structural models (PDB: 4R8P, 7JZV, 7LYB) relative to H2A K119 in the RING1B/BMI1/nucleosome complex (PDB: 4R8P, orange sphere). The Cα-Cα distances from H2A K119 to the respective E2 active sites in the E3/E2/nucleosome complexes (dashed red lines) are reported inside the active site spheres. For the measurements, models were aligned by H2B in the nucleosome. (D, E) BRCA1/BARD1 RING-histone interactions from two published structural models (PDB: 7JZV and 7LYB). The models are aligned by H2B on the E3/E2-bound face of the the nucleosome (see H2B αC helix) and shown side-by-side instead of overlaid for clarity. The largest difference between the two structures is in the positioning of the BARD1 RING domain. Critical histone-binding BRCA1/BARD1 sidechains and the H3 K79 sidechain in the H3 α1-L1 elbow are shown as sticks. Histone colors are indicated in the legend, and BRCA1 is red and BARD1 is blue.
