Extended Data Fig. 7. Phased CTCF-targeted DiMeLo-seq reads.
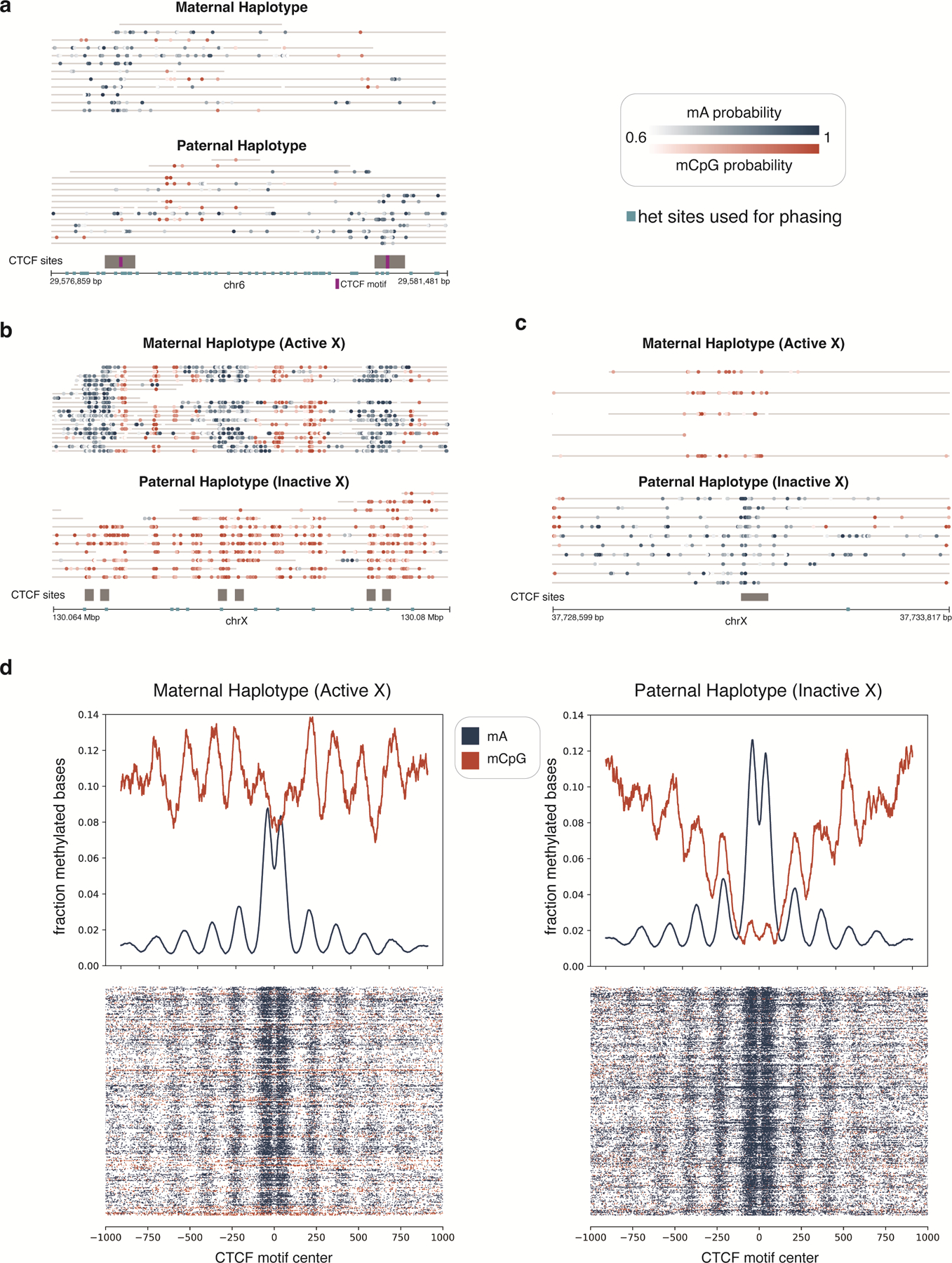
Phased reads across one region on chr6 and two regions on chrX illustrate haplotype-specific CTCF binding due to genetic and epigenetic differences between haplotypes. a, A region on chr6 within the human leukocyte antigen (HLA) locus which contains two CTCF binding sites and many heterozygous SNPs useful for phasing reads. Both CTCF binding sites overlap a het SNP within their binding motif. At the first CTCF site, the paternal SNP allele within the motif is associated with weak or no CTCF binding on the paternal haplotype, and the opposite is true at the second CTCF site. Thus, only one of these two neighboring sites tends to be bound on each haplotype, which is clearly visible on reads spanning both CTCF sites. Further, because CpG methylation patterns are similar between the two haplotypes, these binding differences likely owe to the genetic differences present in/near the CTCF binding motifs themselves. b-c, Because the GM12878 cell line has two X chromosomes and was clonally derived, one X homolog (the paternally inherited X homolog for this cell line) has undergone X inactivation and remains inactive in all cells. Shown here are one region with CTCF binding on the active X only (b) and one region with CTCF binding on the inactive X only (c). The haplotype-specific CTCF binding patterns in these chrX regions appear to be associated with haplotype-specific CpG methylation, as similarly seen for the imprinted H19 locus shown in Fig. 4d. d, Aggregate enrichment profiles from DiMeLo-seq reads across all CTCF sites on chrX are shown, as in Fig. 4b. Each row in the heatmaps below the aggregate plots represents a single molecule centered at the CTCF motif. Notable strips of CpG hypermethylated reads are visible on the active X, as observed previously 12,55.
