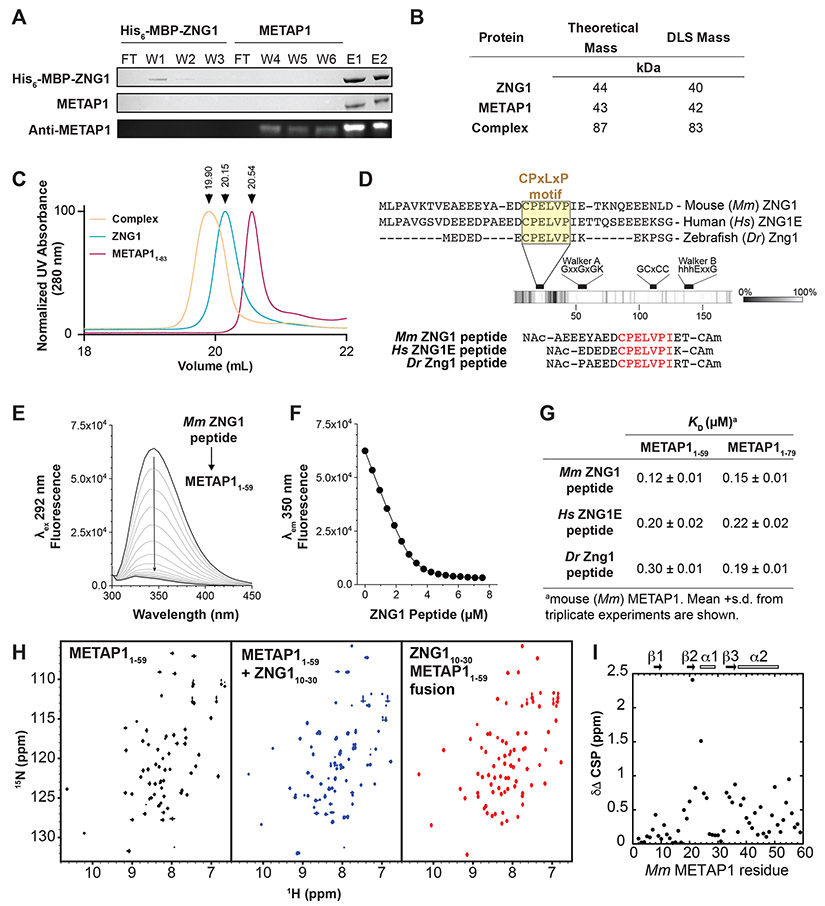
Figure 2. ZNG1 interacts with METAP1 via a unique C6H2 Zn finger domain.
(A) Affinity chromatography of mouse (Mm) His-MBP-ZNG1 bound to amylose resin with purified Mm METAP1. Detection by SDS PAGE (top 2 panels) and immunoblot (bottom). W-wash, F-flow through, E-elution. (B) DLS analysis of full-length Mm ZNG1 and METAP1 shows complex formation in vitro. (C) Size exclusion chromatography of full-length Mm ZNG1 and METAP11-83. (D) Identification of a conserved N-terminal ‘CPELVPI’ motif in Mm, human (Hs), and zebrafish (Dr) ZNG1s and synthetic peptides. (E) Spectra of W45 fluorescence quenching of 3 μM METAP11-59 upon Mm ZNG1 N-terminus binding. (F) Peptide binding curve from (E) fit to a 1:1 binding model. (G) Measured affinities of METAP11-59 and METAP11-79 binding to Mm, Hs, and Dr ZNG1 N-termini. (H) 1H 15N HSQC NMR spectra of METAP11-59 alone (left), in complex with Mm ZNG1 N-terminal peptide (middle), and as N-terminal Mm ZNG1 peptide fusion (right). (I) Chemical shift perturbations in 15N METAP11-59 upon binding to Mm ZNG1 peptide. See also Figure S2.