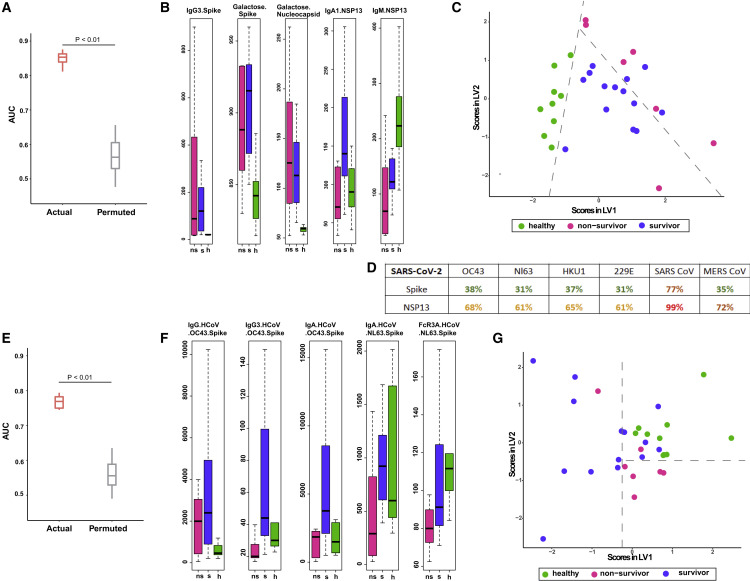
Figure 3.
Antibodies directed against endemic CoV antigens as predictors of severe COVID-19 outcome
(A) Performance of LASSO model to discriminate between survivors, non-survivors, and healthy controls built using deep humoral profiles against canonical and non-canonical Ag specificities. Model performance (three way) is measured in a k-fold cross-validation framework with permutation testing. Actual denotes the performance of the model, built on real data. Permuted denotes performance of the model on shuffled data in a matched cross-validation framework (negative control).
(B) LASSO-selected features from model built using deep humoral profiles against canonical and non-canonical Ag specificities. h, healthy.
(C) PLS-DA using only the LASSO-selected features from the model in (B) to discriminate between healthy controls, survivors, and non-survivors.
(D) Sequence similarities of SARS-CoV-2 spike and NSP13 with corresponding homologs in OC43, NL63, HKU1, 229E, SARS CoV, and MERS CoV.
(E) Performance of LASSO model to discriminate between survivors, non-survivors, and healthy controls built using deep humoral profiles against non-SARS-CoV-2 Ag specificities. Model performance is measured in a k-fold cross-validation framework with permutation testing (negative control) for outcome prediction between survivors, non-survivors, and healthy controls (three way).
(F) LASSO-selected features from model built using deep humoral profiles against non-SARS-CoV-2 Ag specificities.
(G) PLS-DA using only the LASSO-selected features from the model in (F) to discriminate between healthy controls, survivors, and non-survivors.