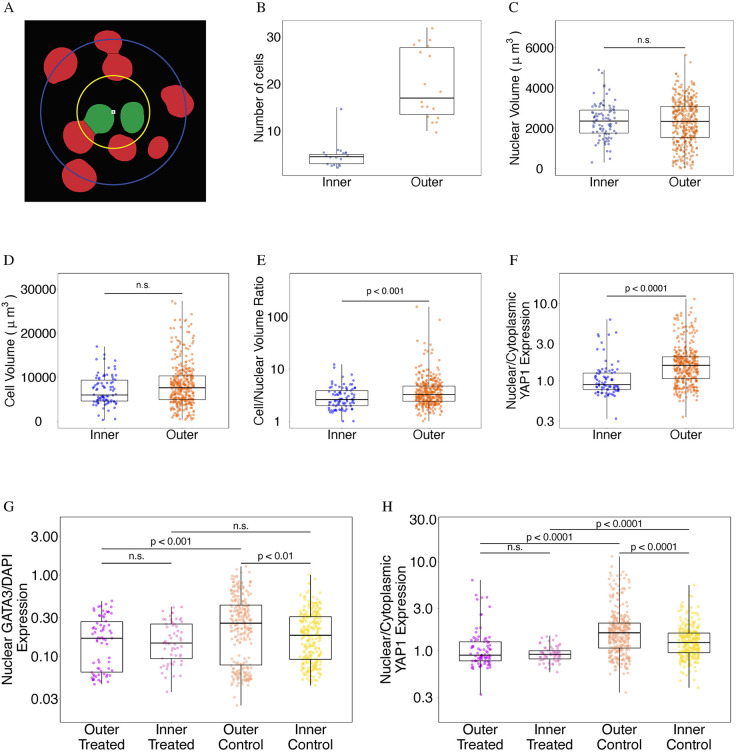
Fig. 4.
GIANI reveals differences in morphology and protein expression in mouse embryos. Unless otherwise stated, each dot represents a single cell. The GIANI settings used to produce this data are available to download from https://doi.org/10.5281/zenodo.6206008. (A) Illustration of the division of embryo cells into ‘outer’ (red) and ‘inner’ (green) subpopulations. The embryo centroid is indicated by the white square. The blue circle has radius Dm and indicates the distance from the embryo centroid to the most distant nucleus centroid. The radius of the yellow circle is DT×Dm. (B) The number of cells in each embryo divided into outer and inner sub-populations using a value of 0.5 for DT in control embryos. Each dot represents a single embryo (ncontrol=18). (C) Volume of nuclei in inner and outer populations in control embryos. (D) Volume of cells in inner and outer populations in control embryos. (E) Ratio of cell-to-nuclear volume in inner and outer cells in control embryos. (F) Nuclear/cytoplasmic ratio of YAP1 expression in control embryos. (G) Difference in expression profiles of nuclear GATA3 expression, normalised to DAPI, in control and treated embryos (ntreated=20). (H) Difference in nuclear/cytoplasmic ratiometric expression profiles of YAP1 in control and treated embryos. Box plots show the median (line) and IQR, with the whiskers extending 1.5 times the IQR from the 25th and 75th percentiles. n.s., not significant (two-sample Wilcoxon tests).