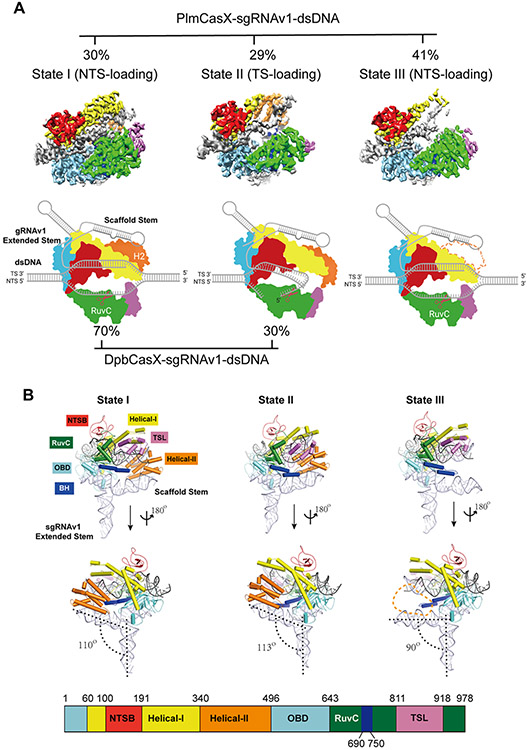
Figure 2. Overall structures of the dPlmCasX-sgRNAv1-dsDNA complex.
(A) The different structural states of the dPlmCasX ternary complex with the sgRNAv1 scaffold revealed by single particle cryo-EM. The top views of refined EM maps for States I, II and III are shown in the top panel. The three maps are shown at contour thresholds of 6 to 9 times sigma. The cartoon model for each map is presented in the bottom panel for better elucidation of substrate DNA loading and cleavage. Referring to the published DpbCasX maps (Liu et al., 2019), the NTSB domain is colored in red, Helical-I in yellow, Helical-II in orange, OBD in aquamarine, RuvC in green, TSL in pink and the bridge helix (BH) in blue. The sgRNAv1 is in light gray and the dsDNA is in dark gray. The invisible Helical-II (H2) domain in State III is represented with a dashed line. The particle proportions for all functional states within the PlmCasX complex (determined in this study) and DpbCasX complex (Liu et al., 2019) are presented with percentages. (B) The atomic models of the dPlmCasX-sgRNAv1-dsDNA complex in three states shown in a front and back view. The domain architecture of the PlmCasX amino acid sequence is shown in the bottom panel. The protein domains in the atomic models share the same color codes as in A. The angle between the sgRNAv1 scaffold stem and extended stem (defined by RNA helix rotation axis, black dashed line) was calculated in PyMol. The Helical-II domain region is outlined with an orange dashed line in State III.