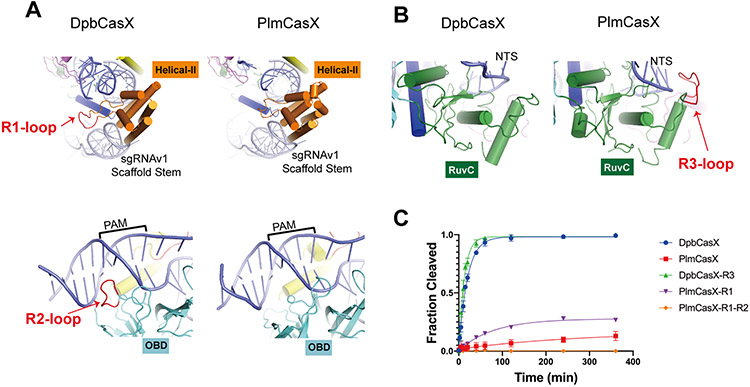
Figure 3. Structural comparison between DpbCasX and PlmCasX.
(A) Region 1 (R1) and region 2 (R2) loops are located within DpbCasX but are absent from PlmCasX. The protein domains are colored as seen in Figure 2; the sgRNAv1 is colored in light gray and the dsDNA (with PAM region labeled) in blue. (B) The region 3 (R3) loop is located within PlmCasX but is absent from DpbCasX. (C) Biochemical dsDNA cleavage activity comparison between CasX chimeras with sgRNAv1 (n = 3, mean ± SD), based on cleavage of the NTS DNA. The rate constant k values for DpbCasX, PlmCasX, DpbCasX-R3 and PlmCasX-R1 were 0.05042, 0.003569, 0.07993 and 0.012503 (fraction/minute), respectively.