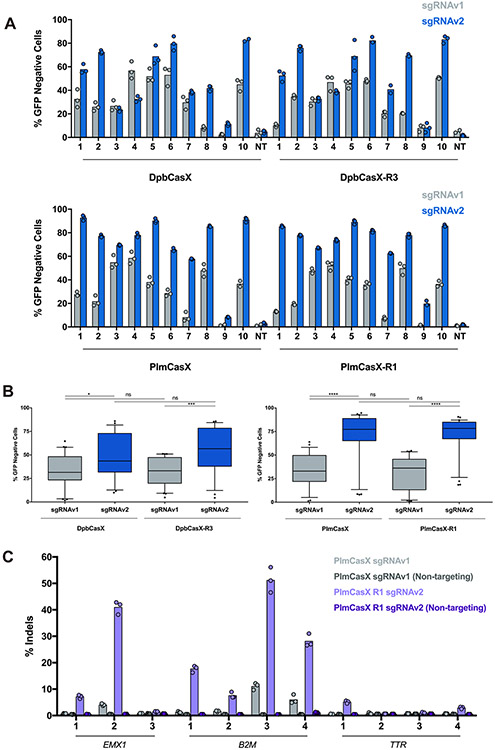
Figure 5. Improved genome editing by engineered DpbCasX and PlmCasX.
(A) Human cell genome editing by DpbCasX, DpbCasX-R3, PlmCasX and PlmCasX-R1 using sgRNAv1 or sgRNAv2 revealed by disruption of genetically encoded GFP. The GFP disruption efficacies for all ten GFP guides are shown (n = 3 (except PlmCasX sgRNAv1 spacers 9, 10, NT; DpbCasX-R3 sgRNAv1 NT; DpbCasX sgRNAv2 spacer 10 and DpbCasX-R3 sgRNAv2 spacer 7; n = 2), mean). NT indicates the non-targeting sgRNA. (B) Genome editing efficacies for all ten GFP-targeting sgRNAs as a box and whisker plot: the box represents the 25th, 50th, and 75th percentile, the whiskers represent the 10th and 90th percentile, and outliers are plotted individually. Significances were determined via one-way ANOVA followed by Tukey’s multiple comparisons test. ns = not significant, * p < 0.05, ** p < 0.01, *** p < 0.001, and **** p < 0.0001. (C) Editing of the human genes EMX1, B2M, and TTR by PlmCasX-v1 or PlmCasX-R1-v2 with multiple spacer sequences in HEK293T cells. The sequences of all spacers are listed in Table S1.