Extended Data Fig. 3. Cryo-EM analysis of Cas9:sgRNA:DNA with 0 RNA:DNA matches.
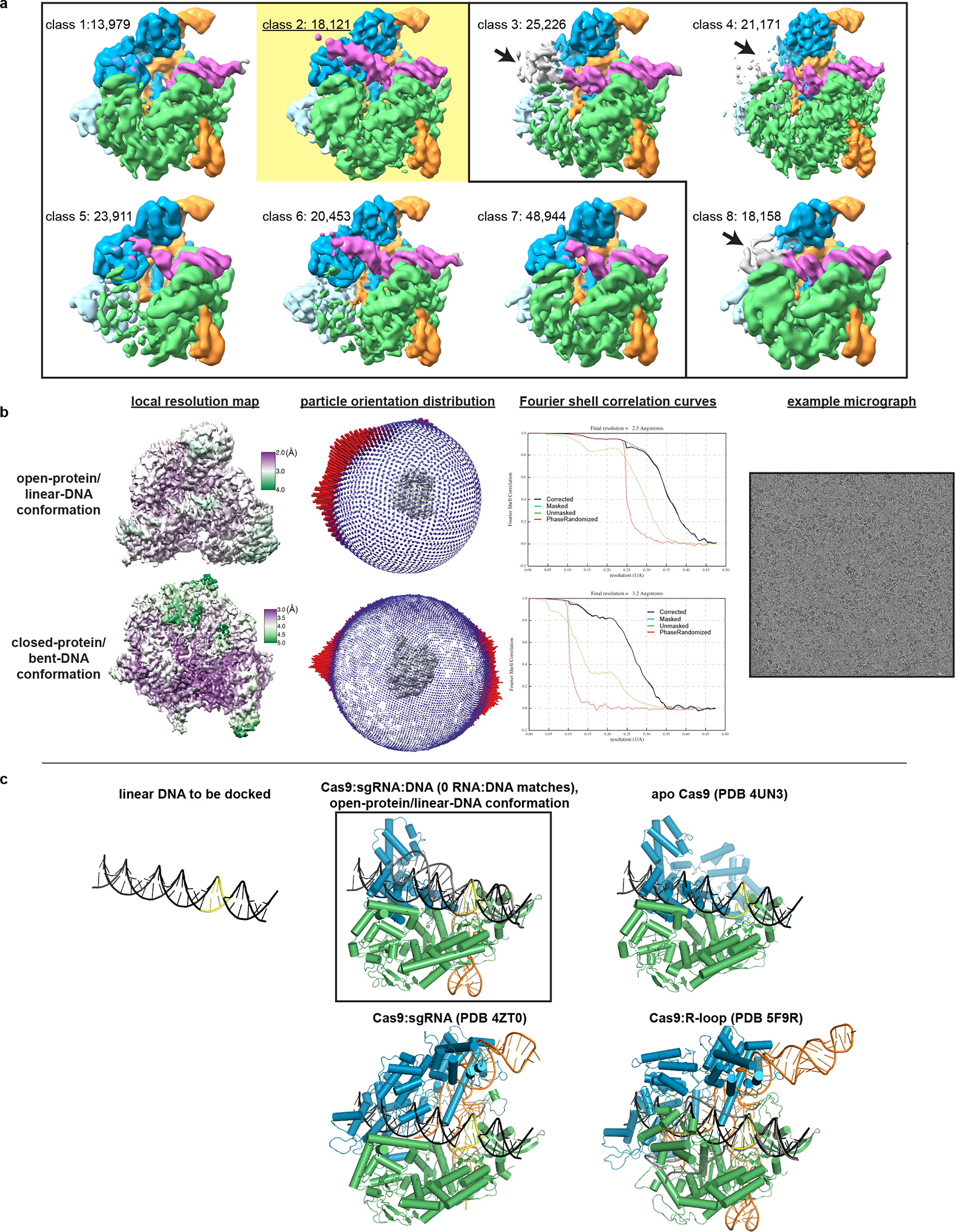
a, Classes from RELION 3D classification of closed-protein particles (threshold 6σ). The number of particles in each class is indicated next to the class number. In classes 1/2/5/6/7 the DNA is bent next to the PAM (visible for class 1 at lower contour). In classes 3/4/8 the DNA continues along a more linear trajectory for half a turn past the PAM, into the region normally occupied by REC2; in these classes, density in the region of the putative collision (black arrow) is uninterpretable as either protein or DNA, likely due to particle damage, and is thus colored gray. The class used for the final closed-protein/bent-DNA map is class 2, highlighted in yellow. Green, NUC lobe; blue, REC lobe domains 1/2; light blue, REC lobe domain 3; orange, guide RNA; magenta, DNA. b, Details of final cryo-EM maps. c, Linear DNA docked into open-protein/linear-DNA cryo-EM structure and previous Cas9 crystal structures. All structures were aligned to the C-terminal domain of PDB 5F9R; then, the linear DNA was aligned to the PAM-containing duplex of PDB 5F9R. Green, NUC lobe; blue, REC lobe; orange, guide RNA; black, docked DNA; yellow, PAM. The DNA truly belonging to each structure (if present) is depicted in gray.
