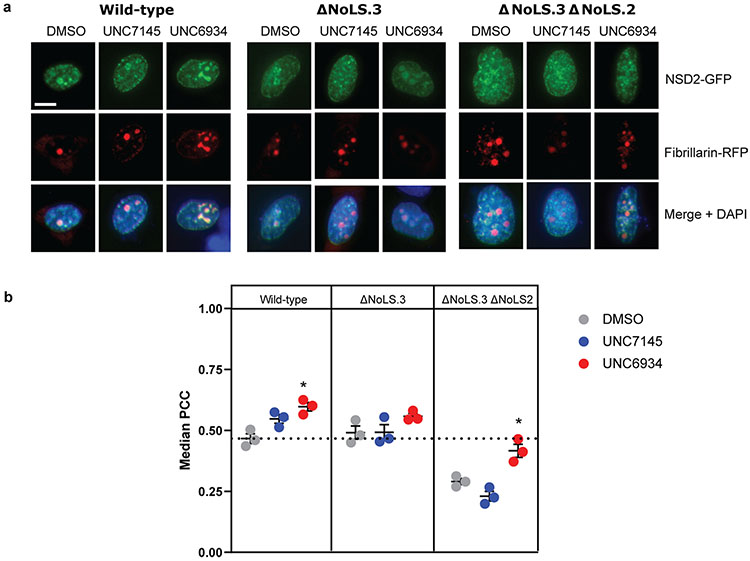
Extended Data Fig. 10 ∣. Deletion of the Nucleolar Localization Sequences significantly reduces NSD2 nucleolar localization driven by UNC6934.
a, Representative confocal microscopy of U2OS cells co-expressing GFP-tagged NSD2 wild-type/mutants and RFP-tagged fibrillarin (scale bar = 10 μm). Transfected cells were treated for four hours with DMSO control, 5 μM UNC6934 or 5 μM UNC7145 and co-localization was measure by Pearson correlation coefficient (PCC). b, Quantification of co-localization between RFP-tagged fibrillarin and NSD2 wild-type or mutants by median PCC per biological replicate between NSD2 and fibrillarin signal across three independent experiments (n = 3; P-values derived from a two-way ANOVA to DMSO control for each panel are indicated as *P = 0.022). For each experiment, PCC/nucleus was measured across ten fields of view. Data shown as mean ± sd.