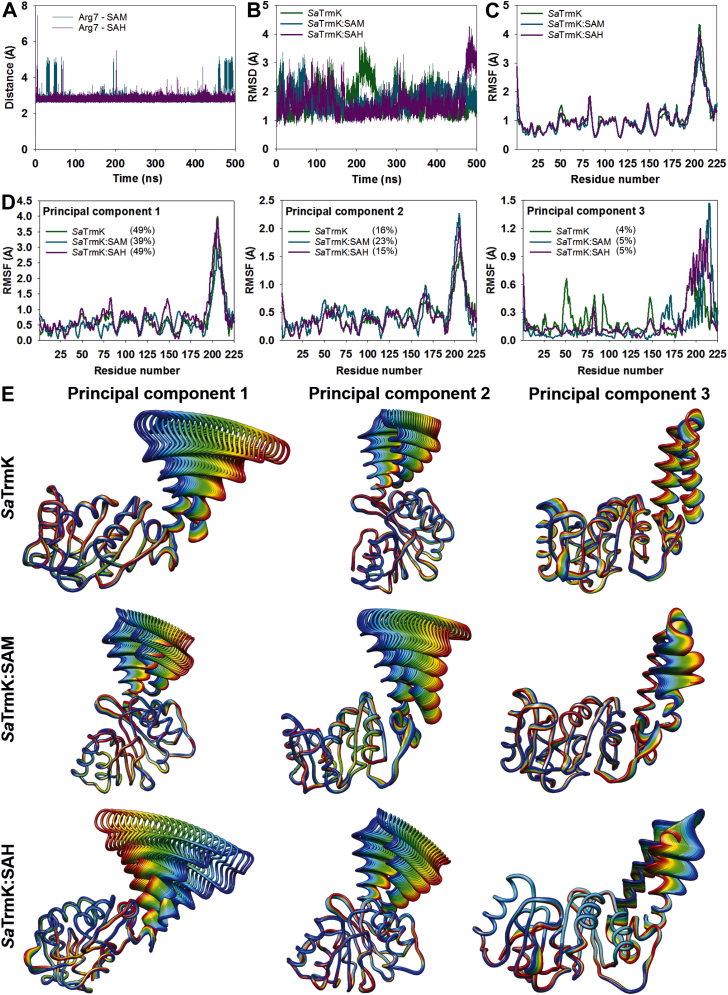
Figure 5.
MD simulations of SaTrmK.A, mean length of the salt bridge between Arg7 and either SAM or SAH. B, time-dependence of the RMSDs over SaTrmK, SaTrmK:SAM, and SaTrmK:SAH Cα. C, mean RMSF over Cα. D, Cα RMSFs of the top three dominant motions of SaTrmK, SaTrmK:SAM, and SaTrmK:SAH as revealed by principal component analysis. Percentages in brackets represent the relative eigenvalue contributed by each eigenvector in each structure. E, ribbon-diagram representation of each dominant motion. The width or delocalization of the ribbon regions corresponds to the motion amplitude. SaTrmK, S. aureus TrmK; TrmK, m1A22-tRNA methyltransferase; MD, molecular dynamics; RMSF, root-mean-square fluctuation.