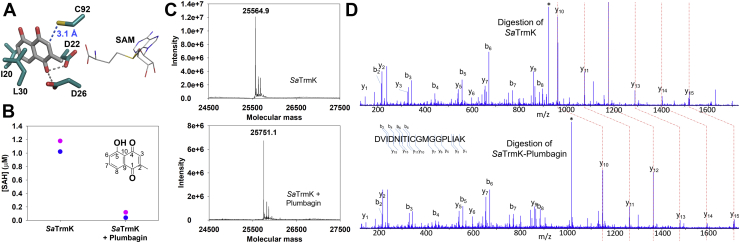
Figure 8.
Inhibition of SaTrmK by plumbagin.A, close-up of the molecular docking-predicted binding of plumbagin to an SaTrmK site adjacent to the SAM-binding site. Plumbagin and protein are shown in stick model, while SAM is shown in wireframe. Oxygen is depicted in red, nitrogen in blue, sulfur in yellow, and carbon in either gray (SAM and plumbagin) or teal (SaTrmK). B, inhibition of SaTrmK by plumbagin. Each data point of the duplicate measurements is shown (pink and blue). The inset depicts the chemical structure of plumbagin with ring carbons numbered. C, intact mass of vehicle-treated (top) and plumbagin-treated (bottom) SaTrmK, showing an increase in molecular mass of 186.2 upon incubation with plumbagin. D, MS/MS fragmentation data for [M+2H]2+ = 923.4972 and [M+2H]2+ = 1016.5033 for trypsin/Glu-C-digested vehicle-treated (top) and plumbagin-treated (bottom) SaTrmK. The b- and y-fragmentation ions are indicated, with shift of 186.0122 at y10. The ∗ denotes the unfragmented peptide. SaTrmK, S. aureus TrmK; TrmK, m1A22-tRNA methyltransferase.