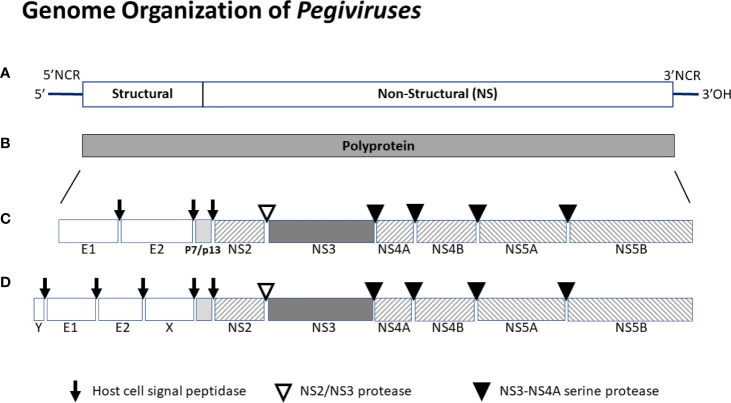
Figure 2.
Schematic of the genome organization of pegiviruses. Pegivirus genomes range from 8.9 to 11.3 kb, and those with longer genomes (e.g. HPgV-2) produce two additional structural proteins (X and Y). The genome (A) has a highly structured 5’ noncoding region (NCR) that contains an internal ribosomal entry site directing translation, and a 3’ NCR. The positive sense viral genome is translated and encodes a single polyprotein (B) that is co- and post-translationally cleaved into the final viral proteins (C or D). Structural proteins are cleaved by host cell signal peptidases. Nonstructural proteins NS2-NS3 are cleaved by the NS2-NS3 autoprotease while NS3 to NS5 proteins are cleaved by the NS3-NS4A protease complex. All pegiviruses have two structural proteins (envelope glycoproteins E1 and E2), and those with longer genomes have two additional predicted structural proteins - a basic protein of unknown function upstream of E1 (Y) and some pegiviruses contain a predicted glycoprotein downstream of E2 (X). The RNA dependent, RNA polymerase resides in NS5B. The human pegivirus (HPgV-1) genome is approximately 9.4 kb and the final protein organization is shown in the middle panel (B).