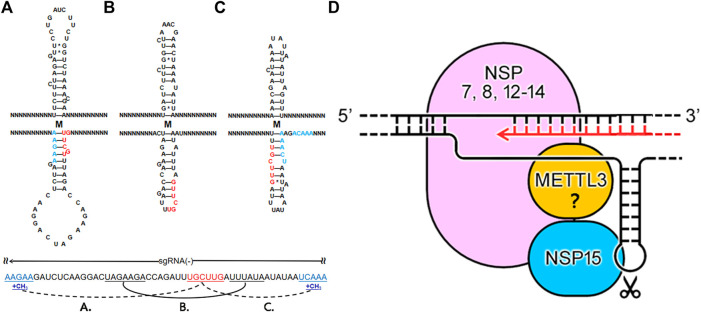
FIGURE 4.
How RTC functions in jumping transcription. N represents any nt. Using the M gene of SARS-CoV-2 as an example, the first (A) and third (C) hairpins require the “AAGAA-like” or AAACH motifs involved in the base pairing. RNA methylation of sequences containing the “AAGAA-like” or AAACH (in blue color) motifs is not in favour of base pairing, ensuring the formation of a unique TRS hairpin (B) containing a NSP15 cleavage site in the loop (D) 5′-3′ represents the strand of the SARS-CoV-2 genome. NSP12-14 form the main structure of the RTC; NSP7 and NSP8, acting as the cofactors of NSP12, may be also included in the main structure of the RTC (Yan et al., 2020); NSP15 and METTL3 are coupled with the main structure. The RTC processes the double-strand RNAs (dsRNAs) and single-strand RNAs (ssRNAs) in two situations. Nascent RNAs are synthesized in one route using unwound ssRNAs(+) or ssRNAs(-) as templates. In the other route, ssRNAs(-) can be uncleaved or cleaved for jumping transcription or degraded, which is regulated by a negative feedback mechanism (Li et al., 2021a). NSP15 cleaves a ssRNA in a small loop in the second route.