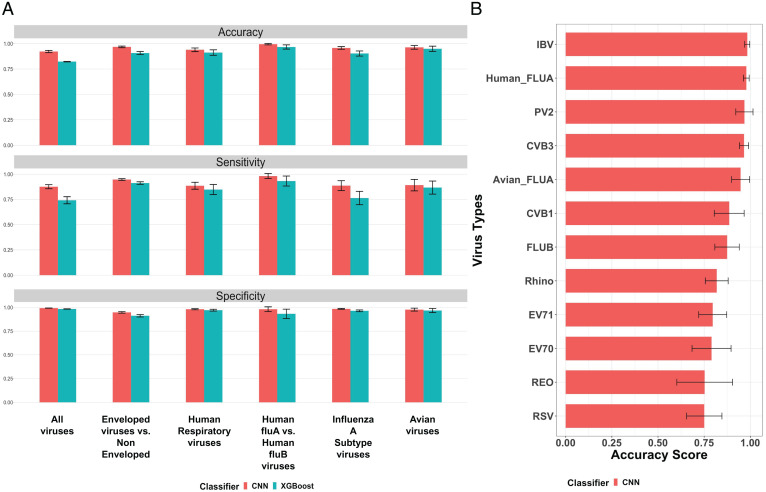
Fig. 4.
(A) The classification performance of our CNN model and the XGBoost model on six experiments: 1) all viruses (classification of all virus types): avian, enteroviruses, human respiratory viruses; 2) enveloped viruses versus nonenveloped: FLUA and FLUB, IBV coronavirus, and RSV are enveloped, and reovirus, enterovirus CVB1/CVB3/EV70/EV71/PV2, and rhino are nonenveloped; 3) human respiratory viruses; 4) human FLUA versus human FLUB viruses; 5) FLUA subtypes; and 6) avian viruses. Three metrics (accuracy, sensitivity, and specificity) are measured for both classification models. Results for all metrics are obtained by running a 5-fold cross-validation five times for fair comparison (each error bar represents the SD of the corresponding metric score for each experiment across 5-fold cross-validation in five tests). (B) Accuracy score for every virus type in the all-viruses classification task (each error bar represents the SD of the corresponding accuracy score for each virus type across 5-fold cross-validation in five tests).