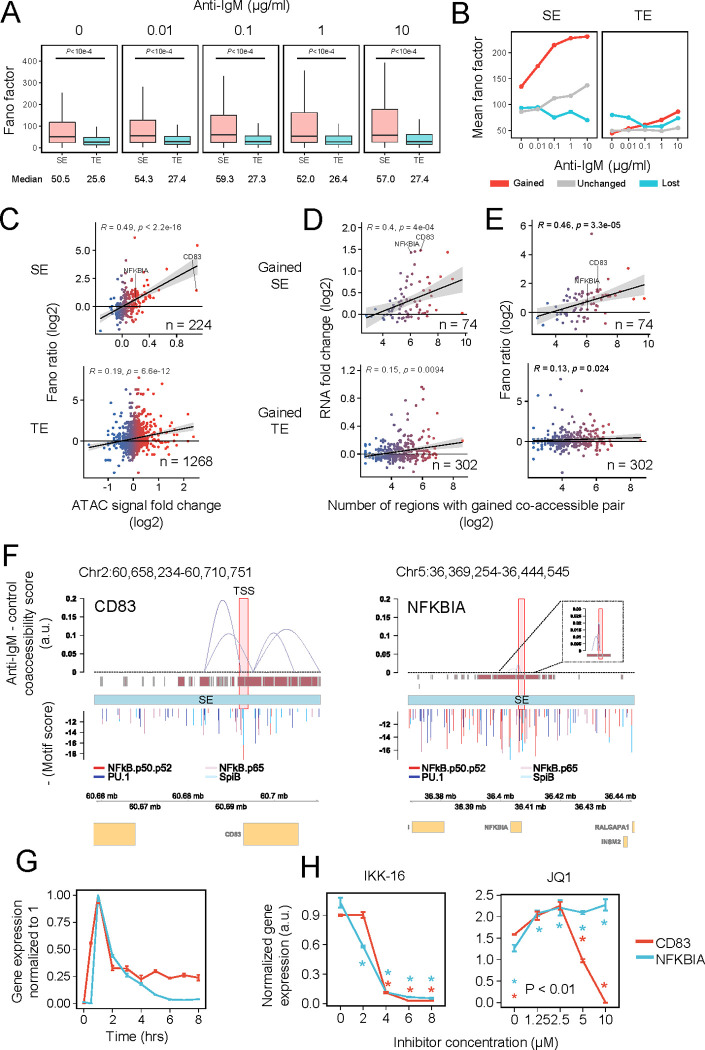
Fig 5. SE relationship with heterogeneity in gene expression.
(A) Boxplot of Fano factor at various doses for DEGs associated with gained, lost, unchanged TE, and SE. Number of genes: SE, 224; TE, 647. P-value was calculated using Welch’s unpaired t-test with undersampling (n = 224). (B) Line plot of mean Fano factor change across dose for SE and TE associated DEGs. Number of genes: Gained SE, 82; Unchanged SE, 100; Lost SE, 42; Gained TE, 242; Unchanged TE, 384; Lost TE, 21. (C) Correlation plot of Fano factor ratios (10 vs. 0 μg/mL anti-IgM-stimulated cells) against ATAC intensity fold changes at SEs and TEs of DEGs. R = Spearman correlation. (D) Correlation plot of RNA fold changes (activated vs inactivated cells) against numbers of co-accessible pairs. (E) Correlation plot of Fano factor ratios (10 vs 0 μg/mL anti-IgM-stimulated cells) against numbers of co-accessible pairs. (F) Co-accessibility score differences obtained using Cicero between stimulated and unstimulated cells shown between genomic regions interacting with regions ±1 kb around the annotated start site of CD83 (left) and NFKBIA (right). (G) Time-course RT-qPCR of CD83 and NFKBIA upon stimulation with 1 μg/mL anti-IgM. Gene expression was normalized to GAPDH. Error bar = SD. (H) RT-qPCR results of IKK-16 (left) and JQ1 (right) treatment 60 min prior to stimulation with 1 μg/mL anti-IgM for 60 min (n = 3). Gene expression was normalized to GAPDH, and P-values were calculated using Student’s unpaired t-test against dose 0 for each dose point. Error bar = SD.