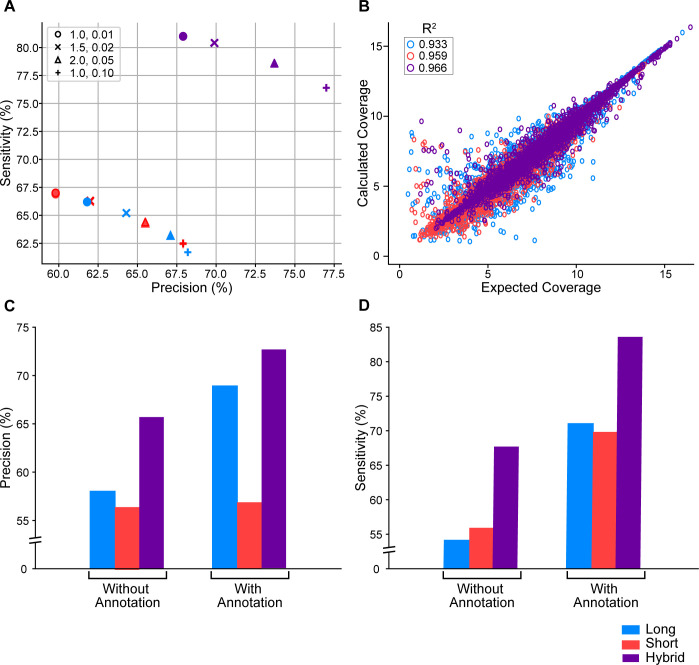
Fig 2.
A) Sensitivity and precision for StringTie assemblies of simulated data with varying sensitivity parameters. The two StringTie parameters varied were the minimum read coverage allowed for a transcript (-c) and the minimum isoform abundance as a fraction of the most abundant transcript at a given locus (-f). Each shape represents a different combination of -c,-f parameters with the values indicated in the legend. The default values of -c and -f are 1.0 and 0.01 respectively and are represented by the circle marker. B) Calculated coverage vs. expected coverage for long-read, short-read, and hybrid-read assemblies of simulated data. Coverage values are normalized to log2(1 + coverage). C) Precision of long-read, short-read, and hybrid-read assemblies of simulated data with and without guide annotation. D) Sensitivity of long-read, short-read, and hybrid-read assemblies of simulated data with and without guide annotation.