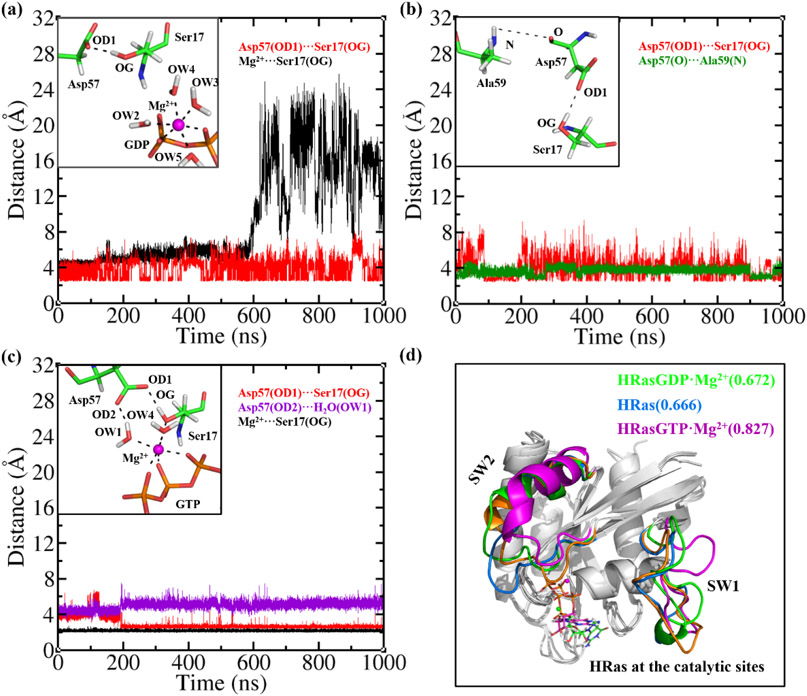
Figure 5.
(a) The distances of OD1-OG and Mg2+-OG at the catalytic site of HRAS·SOS·HRasGDP·Mg2+ during the MD simulations. The inset shows that Ser17 and Asp57 drift away from the Mg2+ ion. (b) The distances of OD1-OG and O-N at the catalytic site of HRAS·SOS·HRas during the MD simulation. The inset shows the H-bond network formed by Ser17, Asp57 and Ala59. (c) The distances of OD1-OG, OD2-OW1 and Mg2+-OG at the catalytic site of HRAS·SOS·HRasGTP·Mg2+ during the MD simulation. The inset shows the stable coordination of Mg2+ to Ser17, Asp57, GTP and the H2O molecules. (d) The least-square fitted conformations of the catalytic HRasGDP·Mg2+, HRas and HRasGTP·Mg2+ in complexes to the reference structure 3L8Z, with the calculated RMSD values of 0.672, 0.666 and 0.827 Å, respectively.